Hello,
I have ran a subject through fmri-prep and have inspected the output files. This subject appears to have an abnormally large sulci. We are waiting on the neuroradiologist’s feedback.
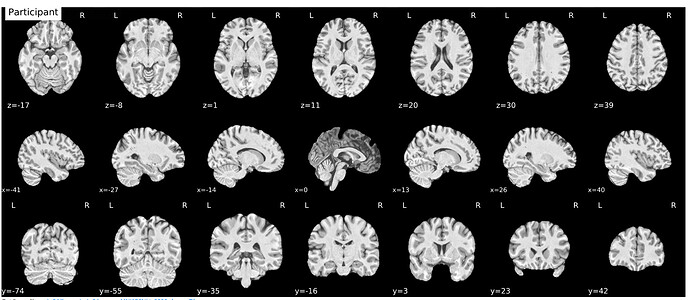
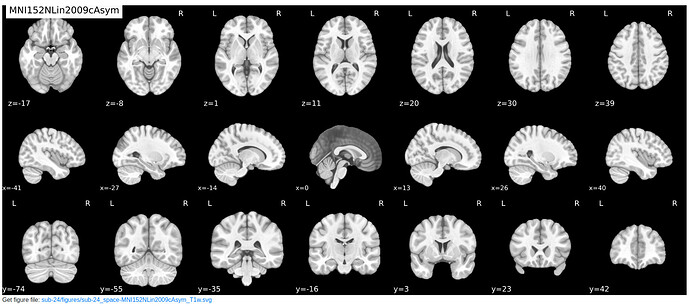
In the meantime, has anyone encountered such a case where the T1w image appears as such (see image: participant at the x=0 coordinate) and the spatial normalization drastically corrects the image to align with the template as such (see image: MNI space)?
Is this a case that would be too extreme in terms of spatial normalization and need to be discarded?
Beware of looking at 3D structures using 2D slices. A feature can appear very thin if cut at a right angle and very wide if cut obliquely. This is the same reason chefs cut carrots at a bias, to increase the surface area for cooking. The x=0 coordinate should run parallel to the longitudinal fissure, so one should expect CSF in this gap. If you view this image with a 3D viewer like MRIcroGL or FSLeyes the true shape of the structure will appear clear.
Therefore, this participant is completely normal and can be included in studies. If you scan a large number of neurologically healthy volunteers, you will occasionally find incidental findings. I have seen people with developmental hydrocephalus and Dandy Walker Malformations. I exclude these individuals from analysis.

1 Like