Hi all. I’ve come across a weird artifact that I’ve not seen before (or seen discussed before) and wanted to see if anyone else had noticed similar things.
Let me caveat this post by saying that I’ve not followed this up super comprehensively, and this artifact may only occur in very specific cases, but I thought it was worth sharing.
Long story short, it appears that transforming a BOLD image to T1w space can induce an odd banding artifact in the data. I suspect this is a byproduct of re-slicing. A (relatively) detailed write-up of the issue can be found here (see the section on “Re-slicing artifacts”), but in summary:
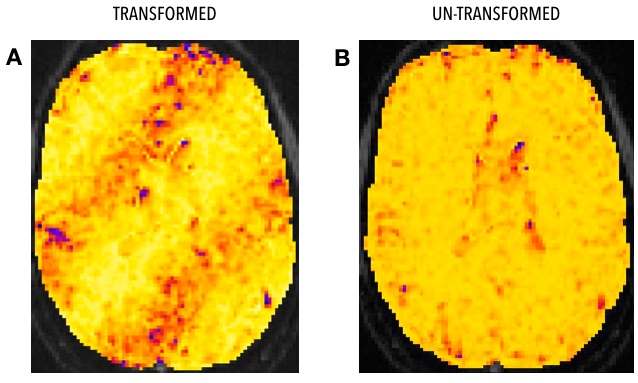
Panel A in the figure above shows the banding artifact in a map of voxel-wise functional connectivity fractional anisotropy (this sounds like a typo but it’s not, see the link above for more info on this weird method), estimated from a BOLD image preprocessed through FMREP and transformed into T1w space. Panel B shows exactly the same thing, but estimated from the un-transformed image.
To chase this down a bit further, I ran MELODIC on the preprocessed data for both transformed and un-transformed images:
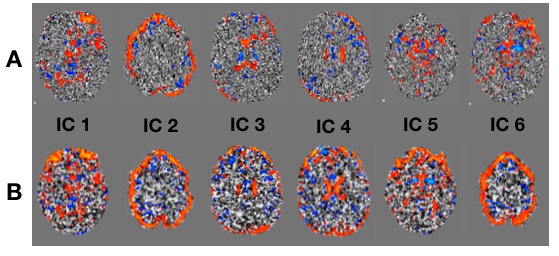
Again, A is transformed data, B is un-transformed data. The diagonal banding is visible in ICs 1-4 and 6 estimated from the transformed data, but is not visible in any of the ICs estimated from the un-transformed data. Perhaps most weirdly, the temporal modes corresponding to ICs 1 and 2 are essentially identical for the transformed and un-transformed data (figures and more detail available in the link above).
Finally, here’s why I think this is caused by re-slicing:
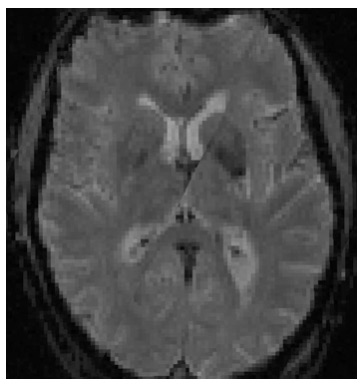
The image above shows how FSLeyes renders the transformed data when displayed alongside the un-transformed data. Closer inspection shows that the diagonal band is essentially the axis along which the most rotation occurs when transforming the image.
Some things that may be driving this:
- This data has relatively anisotropic voxels (1.75 x 1.75 x 3.5mm).
- De-noising regressors included all the standard stuff (e.g. CSF signal, motion parameters) except for WM signal.
I’m not at all clear on how much of an issue this actually is in standard practice (though I have reproduced it on data from multiple individuals). It could well be that this artifact gets removed when regressing out the WM signal, or that it only occurs in edge cases. I plan to chase this up a bit more next month once I have more time, but I wanted to share in the meantime to give a heads up and get thoughts from the community.