Hi all!
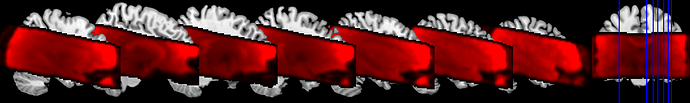
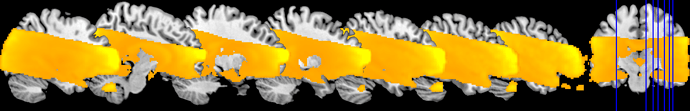
I am working on an odor perception fmri study. I have found some discrepances in BOLD signal between preprocessed data and data after first and second level. Please find attached 2 images with the entire BOLD signal of the FOV.
Preprocessed:
Second Level
Unfortunately, I have no signal in thalamus and globus palidum after the second level analysis.
Does anybody have any ideas what is the reason for this issue?
Thank you!
Best,
Dima
Hey @Dmitriy_Desser,
not 100% sure, but it might be related to SPM’s automatic mask creation during
the estimation of the model. The default threshold for this mask is 0.8, i.e. restricting the estimation only to voxels that exhibit a value that is at least 80% of the global signal present in the included data, hence excluding voxels with an “insufficient” signal over time. If you spend a closer look, the same thing happens around the frontal lobe, that is all regions that are rather dark are excluded. Conversely, the eyes are often included, which is also the case in your data. If you check your data, this might already be present in the 1st level analysis.
My two cents: I like to create brain masks that cover what I need, not more and not less. For example, I’m in auditory neuroscience. Hence, I mostly estimate my models only within the temporal lobe, as I’m not interested in other regions (sorry all you other voxels).
Apparently, you have a slab. You could actually use an approach comparable to what @eknahm and friends did in a part of the Forrest Gump project. They describe very comprehensively how they created a group template based on individual slabs.
Otherwise, you could try creating a mask, that includes every region you are interested in, yourself.
Just drop a message if I/we could be of further help.
HTH, cheers, Peer
Hi,
thank you very much! It works! You have helped me so much!
Cheers,
Dima