Hi SPM experts !
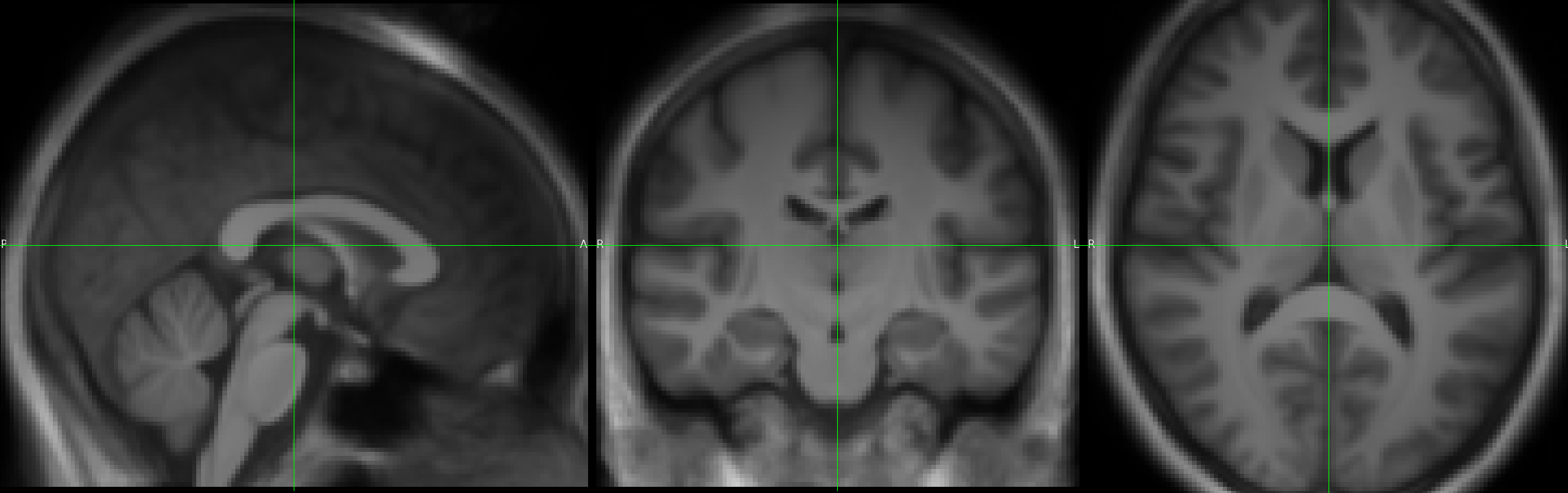
I created a SHOOT template from CAT12 segmentations in ‘affine’ space. I would like to bring 3D and 4D images from the T1 native space to template space. I applied the deformation fields to the original T1s, but the resulting average image is blurry:
I’m probably missing some step or using the wrong settings somehow, would anyone have tips/recommendations on how to do this in a clean way ? I’m not used to SPM functions and I might be missing an obvious solution to this.
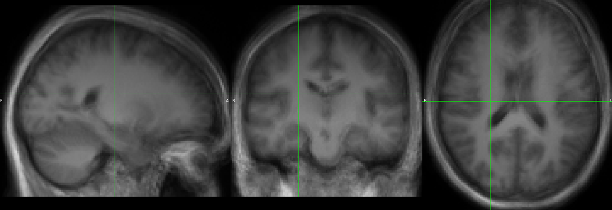
I did manage to tweak CAT12’s code to save the transformation from native to affine space. If I apply this transformation to the T1 images, interpolate them in the same way that CAT12 does, and then apply the deformation field to the T1 images in affine space, the resulting average is crispier:
This is not optimal though, as I’m resampling images in ‘affine’ space (and tweaking CAT12). Is there a way to tell the tool that applies the deformation field about this pre-transform, to avoid intermediate interpolations ? Or change the header of images in native space so that they are in affine space before applying the deformation ?
Additionaly, would be great if this solution could be applied to 4D volumes as well. And even better if there was a way to set the interpolation method. There’s a mask I’d like to interpolate with nearest neighbors, but the tool to apply SHOOT deformation fields doesn’t offer the option to set the interpolation method.
Software Versions:
matlab: R2021b (9.11.0.1769968) 64-bit (glnxa64)
spm12: v7771
CAT12: CAT12.9 (r2577) from 2024-05-03
Code used
I’m using the batch SPM → Tools → SHOOT tools → “Write Normalised” module, with 16 deformation fields generated by the “SHOOT tools → Run Shooting (create Templates)” module (from the CAT12 segmentation files in affine space), to be applied to the 16 native T1 images. I took the voxel size and bounding box values returned when running spm_get_bbox() on the template. I left the template field empty to avoid normalization to MNI space. This led to the blurry image above.
I resampled the images from native space to affine space using something similar to the code below, before applying the deformation field to the image in affine space, by changing the path to the Images to be warped by the SHOOT “Write Normalised” tool. The averaged image looks quite sharper. Unfortunately, I cannot post more than 1 image now (new neurostarts user :p)
for subj_id = {"sub-001", "sub-002", ..., "sub-016"}
load(['/tmp/' subj_id{1} '/mri/transf.mat']); % load transf variable saved from CAT12 segmentation
input_filename = ['/tmp/' subj_id{1} '/' subj_id{1} '_ses-1_T1w.nii'];
[FF, ff, ext] = fileparts(input_filename);
resliced_filename = fullfile(FF, ['r' ff '_affine' ext]);
% Read input info
input_vol = spm_vol(input_filename);
% Save original data
Y = spm_read_vols(input_vol);
% Output vol
output_vol = struct('fname', resliced_filename,'dim',transf.odim,...
'dt', input_vol.dt,...
'pinfo',input_vol.pinfo,...
'mat', transf.mat);
output_vol = spm_create_vol(output_vol);
N = nifti(output_vol.fname);
N.mat = transf.mat;
N.mat0 = transf.mat0;
create(N)
for i=1:transf.odim(3)
tmp = spm_slice_vol(double(Y) ,transf.M*spm_matrix([0 0 i]),transf.odim(1:2),[1,NaN]);
output_vol = spm_write_plane(output_vol,tmp,i);
end
end