Hi all
I am trying to understand the differences between the Small Volume Correctio (SVC) procedure and inclusive masking of the results of a 2nd level GLM analysis using SPM12.
I have 2 groups and have conducted a 2nd level 2-sample t-test using SPM. I proceed with 3 approaches given the fact that I have a strong hypothesis about a specific region of interest. Also, I use 0.05 unc to visualize the results for illustrative purposes.
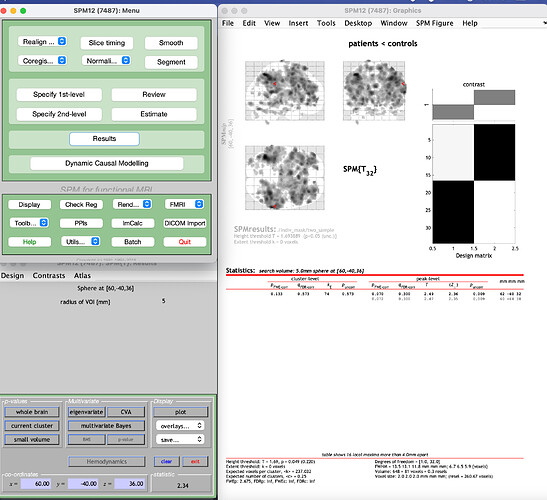
a) I use marsbar to create a sphere at 60 -40 36 with radius 5mm. I save this as .mat using marsbar and then using SPM, I select the “Small Volume” option after visualizing the 2nd level results and then use this .mat as ROI for the SVC analysis.
The results look like this. I get the activity in the whole brain but the results table is limited to the ROI (sphere):
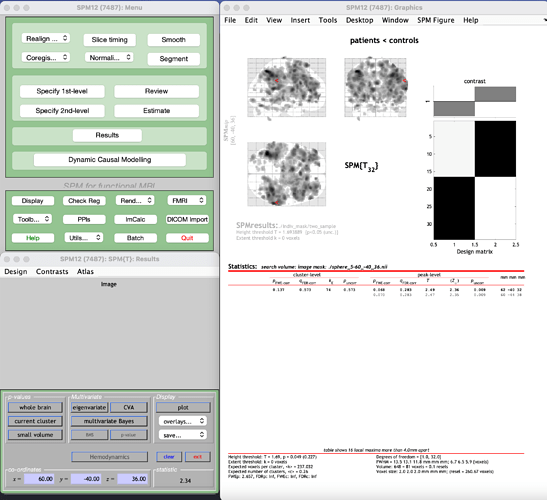
b) I use marsbar to create a sphere at 60 -40 36 with radius 5mm like in a) but now i save is a nifti file in the functional space.
The results look like this but look at the p-values. I see some tiny differences:
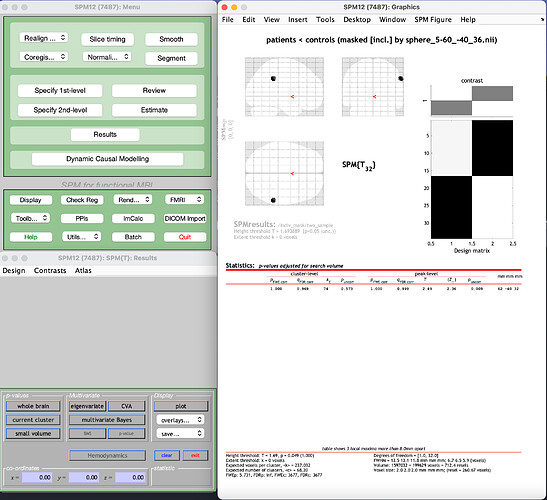
c) finally, after loading the 2nd level results, I do not select “small volume” but instead I choose “apply masking” and then I select my mask image that was created in b).
The results look like this. I get only the activity within the sphere (ROI):
My questions:
-
Why is a) and b) slightly different in terms of reported p-values? It should be identical.
-
How are p-values estimated in a) and b)? Taken into account the whole brain ?
-
In the last case (inclusive masking), how are p-values estimated and what is the main difference between a) / b) and c) ?
Thanks