Hi,
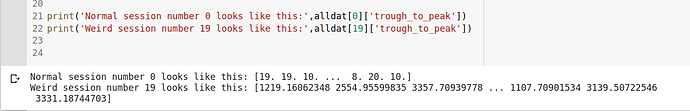
Thanks a lot for the immediate response. We are getting smth like (please see pic).
That doesn’t look right. I checked in the raw data and it’s still there. @nsteinme?
An alternative is to compute widths yourself from the waveforms, which are provided in the “extra” data (see extra notebook).
Weird, yes you’re right, I see that too - about a third of the sessions have nonsense values for waveform duration. I must have had a bug on that (I didn’t actually use it for analyses in the paper, which is why I wouldn’t have caught it). I’m sorry, you’ll have to calculate them from the waveforms provided directly for those sessions.
No I don’t have it easily available, but it is literally just the difference between firing rate prior to trials in the task context and firing rate prior to passive trials, for each neuron.
also to clarify, I think Marius had a typo: 10 samples is 1/3 of a millisecond, not microsecond 
Hi Nick,
Thanks a lot and no worries we will calculate it from the waveform. Thanks for the reply!
Cheers
@pachitarium Hello! Our group is trying to control for retinotopic specificity of neurons in the early visual areas and was wondering if spks_passive also includes recordings during the sparse visual noise stimulus presentation as well as the passive task stimulus replay (auditory tones, gratings…). If yes, could you let us know how to separate them out? If no, would it be possible to have access to the receptive field mapping data?
Thank you so much for your help in advance!
I did not include the retinotopy, I could look into that.
However, I think you have something even better available: whether or not the neurons responded to the contrast_right stimulus. If you can find neurons in multiple areas responsive to stim onset, then you have matched retinotopy. If you want to make sure you’re just looking at a visual response, you can look exclusively at the passive trials.
Generally speaking, the receptive field mapping will probably be noisy and you’ll have trouble estimating the receptive fields and their precise locations. But Nick has targeted the probes to the same retinotopic location based on widefield imaging, so you should be good to go in general.
Hi,
Could you please help with annotation to ‘load steinmetz extra’ data set? Thera are waveforms. What is the duration and amplitude units?
Those are template amplitudes relative to standard deviations of the noise. It’s a little difficult to relate them directly to units of uV, but you can think of them as approximately linearly related. The samples are acquired at 30kHz.
Hi, are LFP still present in your data ? I cannot find them since 2 hours now.
They have been moved to the extra data because they were taking a lot of space. See the new data loader on GitHub for extra data.
Hi,
Could you provide me the exact link to use inorder to get them ? (So I can modify easily my script without problems)?
I use the following script since the beginning:
I use this code to download the data:
fname = []
for j in range(3):
fname.append(‘steinmetz_part%d.npz’%j)
url = [“https://osf.io/agvxh/download”]
url.append(“https://osf.io/uv3mw/download”)
url.append(“https://osf.io/ehmw2/download”)
for j in range(len(url)):
if not os.path.isfile(fname[j]):
try:
r = requests.get(url[j])
except requests.ConnectionError:
print("!!! Failed to download data !!!")
else:
if r.status_code != requests.codes.ok:
print("!!! Failed to download data !!!")
else:
with open(fname[j], "wb") as fid:
fid.write(r.content)
# Data loading
alldat = np.array([])
for j in range(len(fname)):
alldat = np.hstack((alldat, np.load('steinmetz_part%d.npz'%j, allow_pickle=True)['dat']))It’s all in the new data loader script, you’ll have to work it out yourself.
Thank you, but I have this link already.
Even these old ones: https://figshare.com/articles/steinmetz/9598406
https://figshare.com/articles/LFP_data_from_Steinmetz_et_al_2019/9727895
However, what I’m looking for is the new direct link to the LFP data themself (where they have been moved to).
All the information you need about projects is in there, please read the github page carefully. The extra notebook is “Steinmetz extra”:
Thanks so much for the help!
Hi all,
I wanted to ask how one would do PCA on units across all sessions for a specific region and condition eg representation of rewarded vs unrewarded trials in the thalamus.
The way I approached it is the following:
- select the same amount of rewarded and unrewarded trials eg 30 for each recording session
- arrange it into two blocks ([0:30] rewarded, [30:60] unrewarded) to ensure consistency of conditions across sessions (so that each row will have the same condition)
- combine data from different sessions into matrix, where columns are units and rows are responses at specific trial and time
- perform PCA
I was wondering whether it is appropriate to it the way I described. Any comment will be appreciated.
Thanks,
It wouldn’t really help you to combine trials in that way, because the neurons were not recorded simultaneously. It would in fact be better to average across all rewarding and all non-rewarding trials and then concatenate the average timecourses for those two conditions across sessions.