Hi there,
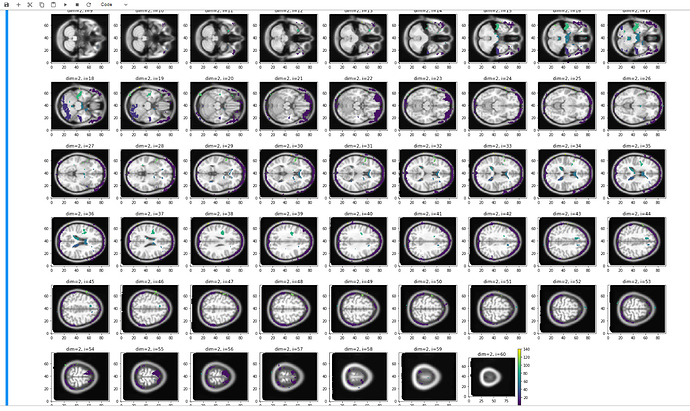
After using fMRIPRep for our 3T mind-wandering study, we performed a custom residual GLM in Nipype (with task and fmriprep-confound regressors) to do seed-based correlation analysis. Our results were as expected for the default mode network, but very strange for the anti-correlated network, showing patterns of activity concentrated around the brain edges and in the ventricles (see image). We did several re-runs of the residuals workflow (with and without confound regressors, with and without smoothing, etc.), which did not change the result.
Finally, we re-run the pipeline with a custom pre-processing workflow instead of fMRIPrep, which did solve the issue. Any ideas how fMRIPrep could have affected these results?
which gave you the result you expected…
BTW - In our paper (https://doi.org/10.1038/s41592-018-0235-4) we show that better preprocessing does not necessarily mean higher t-stats etc.
That looks like you did not use the MNI mask. Also, what MNI template is that you used for this image? Is it MNI152NLin2009cAsym of fMRIPrep?
The first one I can think is that fMRIPrep does not smooth your data. How is that implemented in the alternative pipeline.
Finally, what version of fMRIPrep did you use?.
@oesteban thank you for replying so fast!
The MNI template used for the image is the MNI152NLin2009cAsym, as used in fMRIPrep.
We implemented smoothing the same way in our post-fMRIPrep workflow as we did in our custom preprocessing pipeline, using FSL SUSAN (6mm fwhm).
We used fMRIPrep version 1.1.6.