Summary of what happened:
I am going through the outputs of fmriprep and am noticing that the distortion correction (based on B0 fieldmaps) looks strange. Any insights on what might be happening would be appreciated.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \\
-B /home/hfluhr/shares-hare/ds-learning-habits:/data \\
-B /scratch/hfluhr/workflows/ds-learninghabits:/work \\
-B /home/hfluhr/shares-hare/ds-learning-habits/derivatives:/out \\
/home/hfluhr/data/containers/fmriprep24 /data /out/fmriprep-24.0.1_new participant --participant-label sub-09 -w /work
Version:
24.0.1
Environment (Docker, Singularity / Apptainer, custom installation):
I am running with Singularity.
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.6
[33m1: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)e[39m
./README
[36m Please visit https://neurostars.org/search?q=README_FILE_SMALL for existing conversations about this issue.e[39m
[34me[4mSummary:e[24me[39m e[34me[4mAvailable Tasks:e[24me[39m e[34me[4mAvailable Modalities:e[39me[24m
2203 Files, 50.9GB Reward Pairs MRI
71 - Subjects
1 - Session
[36m If you have any questions, please post on https://neurostars.org/tags/bids.e[39m
Relevant log outputs (up to 20 lines):
240815-14:02:50,644 nipype.workflow IMPORTANT:
fMRIPrep finished successfully!
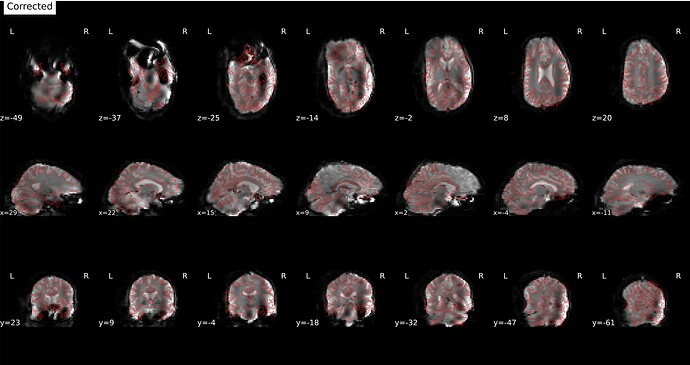
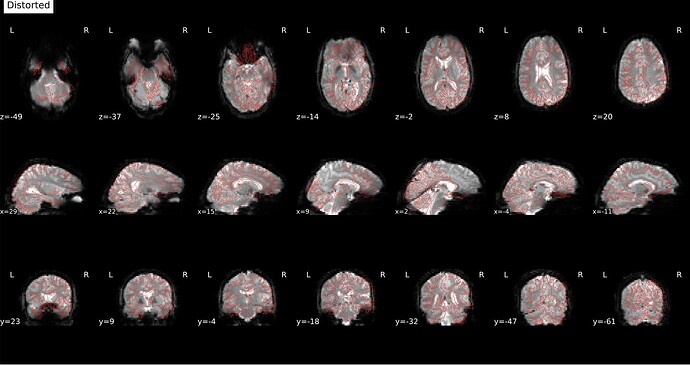
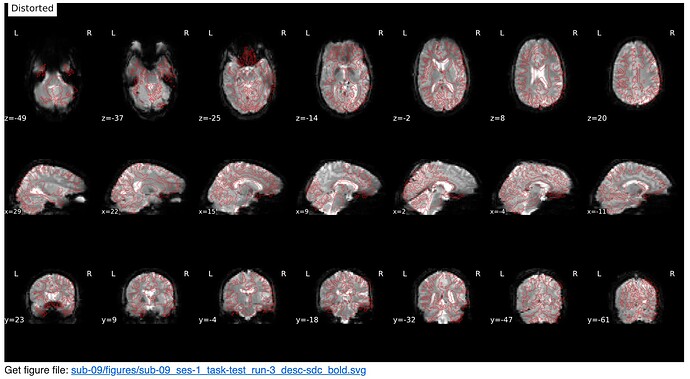
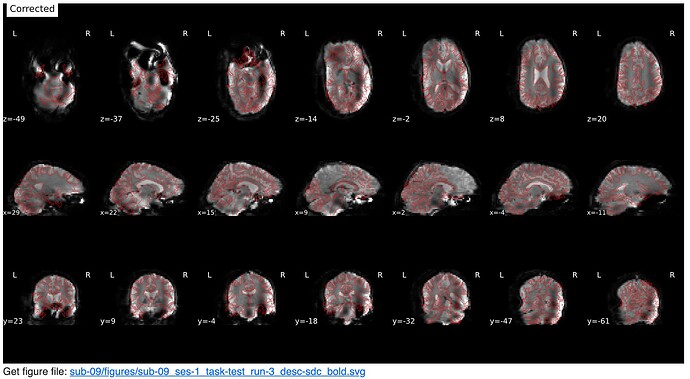
Screenshots / relevant information:
From the html report for a sample subject:
Summary
* Original orientation: LPS
* Repetition time (TR): 2.33s
* Phase-encoding (PE) direction: Right-Left
* Single-echo EPI sequence.
* Slice timing correction: Applied
* Susceptibility distortion correction: FMB (fieldmap-based) - phase-difference map
* Registration: FreeSurfer `bbregister` (boundary-based registration, BBR) - 6 dof
* Non-steady-state volumes: 0
Corrected:
Distorted: