Dear @emdupre ,
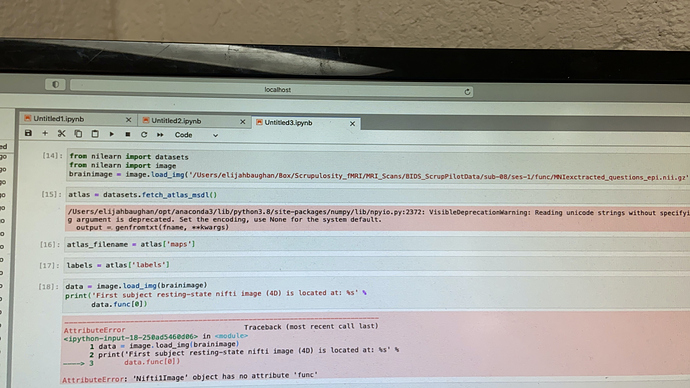
I am trying to perform 9.4.1 “Extracting signals of a probabilistic atlas of functional regions” with some of my own data. The command to bring in the data for the sample data is different than the “image.load_img()” I’m using to bring in my own data. Which I believe is the cause of the attribute errori get, stating that my niftiimage does not have the correct ‘func’ attribute.
Hi @elibaughan !
From that output it looks like you’ve executed image.load_img() one time too many. You could either pass the variable brainimage directly to your print statement or remove the image.load_img() when creating brainimage (so it’s just assigned as a string).
So the relevant code could read:
from nilearn import datasets, image
brainimage = '/path/to/my/image.nii.gz'
data = image.load_img(brainimage)
Hope that helps !
Elizabeth
And maybe one other clarification:
Nibabel objects don’t have a .func attribute; that’s something defined in Nilearn for a dataset. So data.func[0] would be analogous to the path you assigned to brainimage.
An appropriate use of .func would be:
from nilearn import datasets
data = datasets.fetch_development_fmri(n_subjects=3)
print(data.func[0])
@emdupre,
Thank you for the response. It is working now! Sorry for such a simple question, I’m in the process of learning python right now. Regardless, I appreciate your help very much.
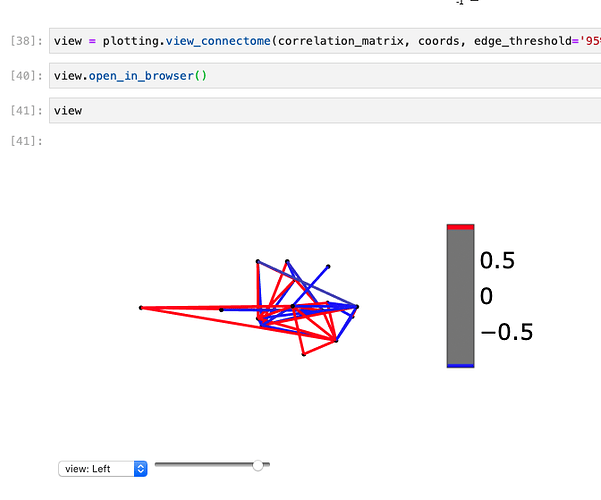
In both the browser form and the Jupiter notebook form of viewing the 3D network (which is so cool!), the brain opacity scale is not working like it does on the example in section 9.4.1.5.
I attached some images below of what it looks like. Any idea of what I may be doing wrong, or something to fix this?
Thank you for help.
Sincerely,
Eli
The brain only appears at full, %100 opacity, hiding the networks. Here is a picture of the opacity almost at 100%, but not showing any brain. Once I scale the opacity to 100%, the brain appears but with no opacity, hiding the network (obviously lol).
Sorry for such a simple question, I’m in the process of learning python right now.
Happy to help ! I know it takes some time to find your way around (especially learning the neuroimaging packages, too !).
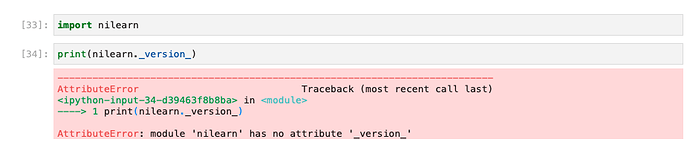
re that output: that does seem wrong. Could you confirm your version of nilearn ? You can use the following:
import nilearn
print(nilearn.__version__)
Other suggestions: Have you tried other browsers? Are you getting any kind of error message when generating or accessing the plot ?
@emdupre,
I tried enter the code you provided but got the following output:
I hope I have nilearn downloaded correctly… lol
However, I did try the html link in Chrome instead of safari, and the opacity worked perfectly!
Thank you yet again.
Sincerely,
Eli
Ah, it should be double underscores (rather than just one) on each side of version. The use of these “dunder” scores is somewhat common in Python; this blog post might provide more useful examples.
If you’re not at version 0.7.0 of Nilearn (the newest release), you can upgrade with
pip install -U nilearn
Regardless, I’m glad that it’s working for you now !
Stay well,
Elizabeth