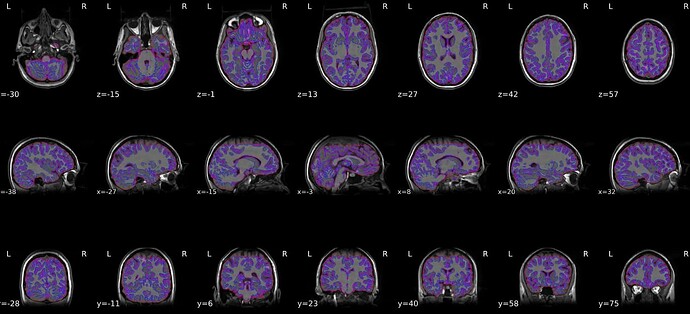
Hi again,
I have given it a go but encountered an error in relation to source_file not being defined.
However I am unsure about how to set the source_file and if the current files are sufficient to carry out the remaining steps of fMRIprep as usual (i.e., just skipping brain extraction and using a pre-defined T1 brain mask)?
The error:
Node Name: fmriprep_24_0_wf.sub_C001_wf.ds_report_about
File: <workdir>/sub-C001/log/20241014-172825_8fbf422f-d8d2-42db-b8a2-512f8e12cf25/crash-20241014-172917-ds_report_about-d7bc2d1d-04b2-4882-a2a8-c884b0ee6ee0.txt
Working Directory: /tmp/fmriprep_24_0_wf/sub_C001_wf/ds_report_about
Inputs:
acquisition:
atlas:
base_directory: ALL_BIDS/Derivative_skull_stripped_processed
ceagent:
check_hdr: True
cohort:
compress:
data_dtype:
datatype: figures
density:
desc: about
direction:
dismiss_entities: ['echo']
echo:
extension:
flip:
fmap:
fmapid:
from:
hemi:
in_file: ['/tmp/fmriprep_24_0_wf/sub_C001_wf/about/report.html']
inv:
label:
meta_dict:
modality:
mode:
model:
mt:
part:
proc:
reconstruction:
recording:
resolution:
roi:
run:
scans:
session:
source_file:
space:
subject:
subset:
suffix:
task:
to:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 722, in _run_command
result = self._interface.run(cwd=outdir, ignore_exception=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 388, in run
self._check_mandatory_inputs()
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 275, in _check_mandatory_inputs
raise ValueError(msg)
ValueError: DerivativesDataSink requires a value for input 'source_file'. For a list of required inputs, see DerivativesDataSink.help()
When creating this crashfile, the results file corresponding
to the node could not be found.
The command:
apptainer run -B "${myproject}":"${myproject}" --cleanenv "${myproject}/ALL_BIDS/fmriprep-24.0.1.simg" --skip-bids-validation "${myproject}/ALL_BIDS/data_BIDS" "${myproject}/ALL_BIDS/Derivative_skull_stripped_processed" --low-mem participant --participant-label "${SUBJECT}" --fs-license-file "${myproject}/ALL_BIDS/license.txt" --output-spaces MNI152NLin6Asym:res-2 MNI152NLin2009cAsym:res-2 --stop-on-first-crash --cifti-output --use-syn-sdc --bids-filter-file "${myproject}/ALL_BIDS/data_BIDS/config.json" --skull-strip-t1 skip --derivatives "${myproject}/ALL_BIDS/Derivative_skull_stripped_processed/${SUBJECT}/anat/" -w /tmp/
The config.json file
{
"t1w": {
"datatype": "anat",
"acquisition": null,
"reconstruction": "rec-optiBET02",
"suffix": "T1w"
}
}
Below are the content of the subject input and output folder before fMRIprep:
The input subject directory
data_BIDS
└── sub-C001
├── anat
│ ├── sub-C001_rec-optiBET02_T1w.json
│ └── sub-C001_rec-optiBET02_T1w.nii.gz. (this is optiBET skull-stripped T1w)
├── func
├── sub-C001_task-rest_bold.json
└── sub-C001_task-rest_bold.nii.gz
The output subject directory
Derivative_skull_stripped_processed
└── sub-C001
├── anat
├── sub-C001_rec-optiBET_desc-brain_mask.json
└── sub-C001_rec-optiBET_desc-brain_mask.nii.gz. (this is the brain mask generated from optiBET)
Content of the sub-C001_rec-optiBET_desc-brain_mask.json
{
"RawSources": [
"ALL_BIDS/data_BIDS/sub-C001/anat/sub-C001_rec-optiBET02_T1w.nii.gz"
],
"Type": "Brain"
}
The output log
subject number: sub-C001
241014-17:28:37,690 nipype.workflow IMPORTANT:
Running fMRIPrep version 24.0.1
License NOTICE ##################################################
fMRIPrep 24.0.1
Copyright The NiPreps Developers.
This product includes software developed by
the NiPreps Community (https://nipreps.org/).
Portions of this software were developed at the Department of
Psychology at Stanford University, Stanford, CA, US.
This software is also distributed as a Docker container image.
The bootstrapping file for the image ("Dockerfile") is licensed
under the MIT License.
This software may be distributed through an add-on package called
"Docker Wrapper" that is under the BSD 3-clause License.
#################################################################
241014-17:28:37,806 nipype.workflow IMPORTANT:
Building fMRIPrep's workflow:
* BIDS dataset path: data_BIDS.
* Participant list: ['C001'].
* Run identifier: 20241014-172825_8fbf422f-d8d2-42db-b8a2-512f8e12cf25.
* Output spaces: MNI152NLin6Asym:res-2 MNI152NLin2009cAsym:res-2.
* Searching for derivatives: [PosixPath('Derivative_skull_stripped_processed/sub-C001/anat')].
* Pre-run FreeSurfer's SUBJECTS_DIR: Derivative_skull_stripped_processed/sourcedata/freesurfer.
241014-17:28:38,771 nipype.workflow INFO:
ANAT Stage 1: Adding template workflow
241014-17:28:39,159 nipype.workflow INFO:
ANAT Found brain mask
241014-17:28:39,159 nipype.workflow INFO:
ANAT Skipping skull-strip, INU-correction only
241014-17:28:39,172 nipype.workflow INFO:
ANAT Stage 3: Preparing segmentation workflow
241014-17:28:39,177 nipype.workflow INFO:
ANAT Stage 4: Preparing normalization workflow for ['MNI152NLin6Asym', 'MNI152NLin2009cAsym']
241014-17:28:39,187 nipype.workflow INFO:
ANAT Stage 5: Preparing surface reconstruction workflow
241014-17:28:39,202 nipype.workflow INFO:
ANAT Found brain mask - skipping Stage 6
241014-17:28:39,202 nipype.workflow INFO:
ANAT No T2w images provided - skipping Stage 7
241014-17:28:39,202 nipype.workflow INFO:
ANAT Stage 8: Creating GIFTI surfaces for ['white', 'pial', 'midthickness', 'sphere_reg', 'sphere']
241014-17:28:39,219 nipype.workflow INFO:
ANAT Stage 8: Creating GIFTI metrics for ['thickness', 'sulc']
241014-17:28:39,225 nipype.workflow INFO:
ANAT Stage 8a: Creating cortical ribbon mask
241014-17:28:39,228 nipype.workflow INFO:
ANAT Stage 9: Creating fsLR registration sphere
241014-17:28:39,232 nipype.workflow INFO:
ANAT Stage 10: Creating MSM-Sulc registration sphere
241014-17:28:39,497 nipype.workflow INFO:
B0 field inhomogeneity map will be estimated with the following 1 estimator(s): [<EstimatorType.ANAT: 5>].
241014-17:28:39,534 nipype.workflow INFO:
Setting-up fieldmap "auto_00000" (EstimatorType.ANAT) with <sub-C001_rec-optiBET02_T1w.nii.gz, sub-C001_task-rest_bold.nii.gz>
241014-17:28:39,703 nipype.workflow INFO:
No single-band-reference found for sub-C001_task-rest_bold.nii.gz.
241014-17:28:39,766 nipype.workflow INFO:
Stage 1: Adding HMC boldref workflow
241014-17:28:39,771 nipype.workflow INFO:
Stage 2: Adding motion correction workflow
241014-17:28:39,778 nipype.workflow INFO:
Stage 3: Adding coregistration boldref workflow
241014-17:28:39,834 nipype.workflow IMPORTANT:
BOLD series will be slice-timing corrected to an offset of 0.972s.
241014-17:28:43,27 nipype.workflow INFO:
fMRIPrep workflow graph with 549 nodes built successfully.
241014-17:28:55,906 nipype.workflow IMPORTANT:
fMRIPrep started!
241014-17:29:11,515 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.syn_preprocessing_auto_00000.anat_dilmsk" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/syn_preprocessing_auto_00000/anat_dilmsk".
241014-17:29:11,516 nipype.workflow INFO:
[Node] Executing "anat_dilmsk" <sdcflows.interfaces.brainmask.BinaryDilation>
241014-17:29:13,296 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.fmap_preproc_wf.out_merge_fmap_id" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/fmap_preproc_wf/out_merge_fmap_id".
241014-17:29:13,297 nipype.workflow INFO:
[Node] Executing "out_merge_fmap_id" <nipype.interfaces.utility.base.Merge>
241014-17:29:13,302 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.fmap_preproc_wf.out_merge_method" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/fmap_preproc_wf/out_merge_method".
241014-17:29:13,303 nipype.workflow INFO:
[Node] Executing "out_merge_method" <nipype.interfaces.utility.base.Merge>
241014-17:29:13,311 nipype.workflow INFO:
[Node] Finished "out_merge_fmap_id", elapsed time 0.000199s.
241014-17:29:13,325 nipype.workflow INFO:
[Node] Finished "out_merge_method", elapsed time 0.000177s.
241014-17:29:13,519 nipype.workflow INFO:
[Node] Setting-up "_validate_nii0" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/syn_preprocessing_auto_00000/epi_reference_wf/validate_nii/mapflow/_validate_nii0".
241014-17:29:13,520 nipype.workflow INFO:
[Node] Executing "_validate_nii0" <niworkflows.interfaces.header.ValidateImage>
241014-17:29:13,684 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.hmc_boldref_wf.get_dummy" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/hmc_boldref_wf/get_dummy".
241014-17:29:13,704 nipype.workflow INFO:
[Node] Executing "get_dummy" <niworkflows.interfaces.bold.NonsteadyStatesDetector>
241014-17:29:13,727 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.hmc_boldref_wf.val_bold" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/hmc_boldref_wf/val_bold".
241014-17:29:13,728 nipype.workflow INFO:
[Node] Executing "val_bold" <niworkflows.interfaces.header.ValidateImage>
241014-17:29:13,780 nipype.workflow INFO:
[Node] Finished "_validate_nii0", elapsed time 0.241188s.
241014-17:29:13,856 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_native_wf.bold_source" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_native_wf/bold_source".
241014-17:29:13,867 nipype.workflow INFO:
[Node] Executing "bold_source" <nipype.interfaces.utility.base.Select>
241014-17:29:13,893 nipype.workflow INFO:
[Node] Finished "bold_source", elapsed time 0.000253s.
241014-17:29:13,908 nipype.workflow INFO:
[Node] Finished "val_bold", elapsed time 0.179408s.
241014-17:29:15,18 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.ds_boldmask_wf.sources" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/ds_boldmask_wf/sources".
241014-17:29:15,19 nipype.workflow INFO:
[Node] Executing "sources" <fmriprep.interfaces.bids.BIDSURI>
241014-17:29:15,20 nipype.workflow INFO:
[Node] Finished "sources", elapsed time 0.000174s.
241014-17:29:15,389 nipype.workflow INFO:
[Node] Finished "get_dummy", elapsed time 1.683674s.
241014-17:29:15,884 nipype.workflow INFO:
[Node] Setting-up "_select_volumes0" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/syn_preprocessing_auto_00000/epi_reference_wf/select_volumes/mapflow/_select_volumes0".
241014-17:29:15,885 nipype.workflow INFO:
[Node] Executing "_select_volumes0" <niworkflows.interfaces.bold.NonsteadyStatesDetector>
241014-17:29:16,171 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.ds_fmapreg_wf.sources" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/ds_fmapreg_wf/sources".
241014-17:29:16,173 nipype.workflow INFO:
[Node] Executing "sources" <fmriprep.interfaces.bids.BIDSURI>
241014-17:29:16,187 nipype.workflow INFO:
[Node] Finished "sources", elapsed time 0.000193s.
241014-17:29:16,247 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_native_wf.validate_bold" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_native_wf/validate_bold".
241014-17:29:16,249 nipype.workflow INFO:
[Node] Executing "validate_bold" <niworkflows.interfaces.header.ValidateImage>
241014-17:29:16,336 nipype.workflow INFO:
[Node] Finished "validate_bold", elapsed time 0.086545s.
241014-17:29:16,519 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.ds_hmc_boldref_wf.sources" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/ds_hmc_boldref_wf/sources".
241014-17:29:16,520 nipype.workflow INFO:
[Node] Executing "sources" <fmriprep.interfaces.bids.BIDSURI>
241014-17:29:16,532 nipype.workflow INFO:
[Node] Finished "sources", elapsed time 0.000247s.
241014-17:29:16,623 nipype.workflow INFO:
[Node] Finished "_select_volumes0", elapsed time 0.699428s.
241014-17:29:16,802 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.select_MNI6_tpl" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/select_MNI6_tpl".
241014-17:29:16,804 nipype.workflow INFO:
[Node] Executing "select_MNI6_tpl" <smriprep.interfaces.templateflow.TemplateFlowSelect>
241014-17:29:16,809 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bidssrc" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bidssrc".
241014-17:29:16,811 nipype.workflow INFO:
[Node] Executing "bidssrc" <niworkflows.interfaces.bids.BIDSDataGrabber>
241014-17:29:16,826 nipype.interface INFO:
No "t2w" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "flair" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "fmap" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "sbref" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "roi" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "pet" images found for sub-C001
241014-17:29:16,827 nipype.interface INFO:
No "asl" images found for sub-C001
241014-17:29:16,827 nipype.workflow INFO:
[Node] Finished "bidssrc", elapsed time 0.000738s.
241014-17:29:16,848 nipype.workflow INFO:
[Node] Finished "select_MNI6_tpl", elapsed time 0.035756s.
241014-17:29:17,63 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.bold_task_rest_wf.bold_fit_wf.ds_hmc_wf.sources" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/bold_task_rest_wf/bold_fit_wf/ds_hmc_wf/sources".
241014-17:29:17,65 nipype.workflow INFO:
[Node] Executing "sources" <fmriprep.interfaces.bids.BIDSURI>
241014-17:29:17,67 nipype.workflow INFO:
[Node] Finished "sources", elapsed time 0.000184s.
241014-17:29:17,439 nipype.workflow WARNING:
[Node] Error on "fmriprep_24_0_wf.sub_C001_wf.ds_report_about" (/tmp/fmriprep_24_0_wf/sub_C001_wf/ds_report_about)
241014-17:29:17,441 nipype.workflow INFO:
[Node] Setting-up "fmriprep_24_0_wf.sub_C001_wf.anat_fit_wf.anat_template_wf.anat_ref_dimensions" in "/tmp/fmriprep_24_0_wf/sub_C001_wf/anat_fit_wf/anat_template_wf/anat_ref_dimensions".
241014-17:29:17,441 nipype.workflow ERROR:
Node ds_report_about failed to run on host spartan-bm021.hpc.unimelb.edu.au.
241014-17:29:17,445 nipype.workflow ERROR:
Saving crash info to Derivative_skull_stripped_processed/sub-C001/log/20241014-172825_8fbf422f-d8d2-42db-b8a2-512f8e12cf25/crash-20241014-172917-ds_report_about-d7bc2d1d-04b2-4882-a2a8-c884b0ee6ee0.txt
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 722, in _run_command
result = self._interface.run(cwd=outdir, ignore_exception=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 388, in run
self._check_mandatory_inputs()
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 275, in _check_mandatory_inputs
raise ValueError(msg)
ValueError: DerivativesDataSink requires a value for input 'source_file'. For a list of required inputs, see DerivativesDataSink.help()
When creating this crashfile, the results file corresponding
to the node could not be found.
241014-17:29:17,482 nipype.workflow CRITICAL:
fMRIPrep failed: Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 722, in _run_command
result = self._interface.run(cwd=outdir, ignore_exception=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 388, in run
self._check_mandatory_inputs()
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 275, in _check_mandatory_inputs
raise ValueError(msg)
ValueError: DerivativesDataSink requires a value for input 'source_file'. For a list of required inputs, see DerivativesDataSink.help()
When creating this crashfile, the results file corresponding
to the node could not be found.
Let me know if any further information is needed and many thanks in advance for your guidance!