Hello experts,
I want to linearly register T1 to DTI with FLIRT, but no matter what rigid-body or affine registeration, there is always a bad alignment, especially in the prefrontal lobe and the posterior part of brain.I tried several ways to solve this error, but I failed again and again. I quite need your help!!!
Here is my code:
- flirt -in brain.nii.gz -ref mean_b0.nii.gz -out t12b0.nii.gz -omat t12b0.mat -dof 6 -cost mutualinfo
*brain.nii.gz is preprocessed by Freesurfer, it is a brain-extracted, bias-field-corrected T1 scan.
*mean_b0.nii.gz is extracted from preprocessed dti data by Mrtrix3, including noise removal, eddy correction, and bias field correction. I only have b-1000 and one PA direction.
**The reason why I do this step is for final registeration of atlas and dti, which is achieved via T1. **
Detailed information is as follow:
- Register T1 to MNI152 template linearly and nonlearly with FLIRT and FNIRT, then I acquire T1-to-MNI152 warping transformation.
- Compute the MNI152-to-T1 warping transformation by inverting the T1-to-MNI152 warping transformation, using FSL’s invwarp command.
- Register the atlas from MNI152 space to T1 space using FSL’s applywarp command, by applying the non-linear MNI152-toT1 warping transformation.
- Register the atlas from T1 space to diffusion space using FSL’s applywarp command.
applywarp --ref=mean_b0.nii.gz --in=atlas --warp=warps_mni2t1.nii.gz --postmat=t12b0.mat --out=atlas_in_dti_space.nii.gz --interp=nn
By visualizing the output data, I found the image quality of the atlas registered to T1 (output at step 4) was good, but the step 5 showed bad output as step 1. So I guess the error is originated from the step 1, wrong t1-to-b0 matrix leads to wrong atlas-to-b0 registeration.
Here are the outputs of step1 and step5
output1: mean_b0.nii.gz (grey color) is overlaid by t12b0.nii.gz (red color), it seems two images are not aligned well.
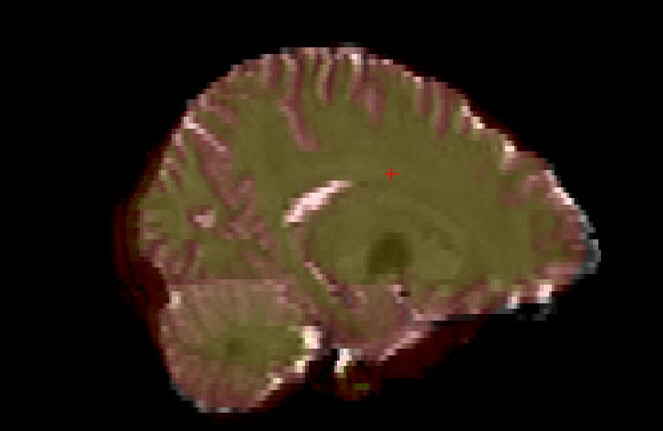
output5: mean_b0.nii.gz (grey color) is overlaid by atals.nii.gz (colorful image)
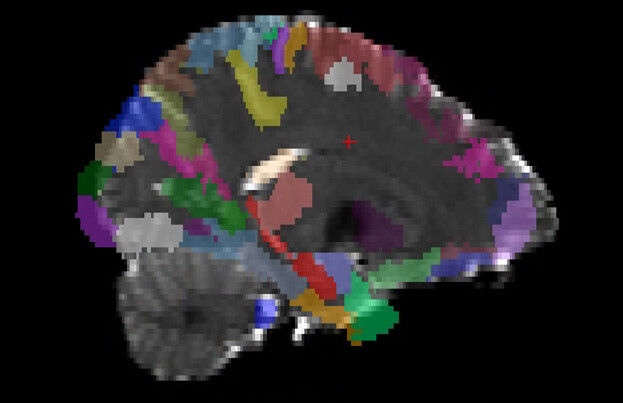
Thank you in advance for any insights!
Best,
Regard