Hi everyone,
I saw similar issue here but the fix suggested did not help so I am posting again.
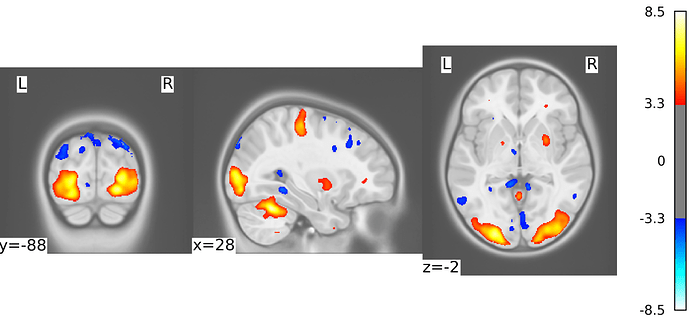
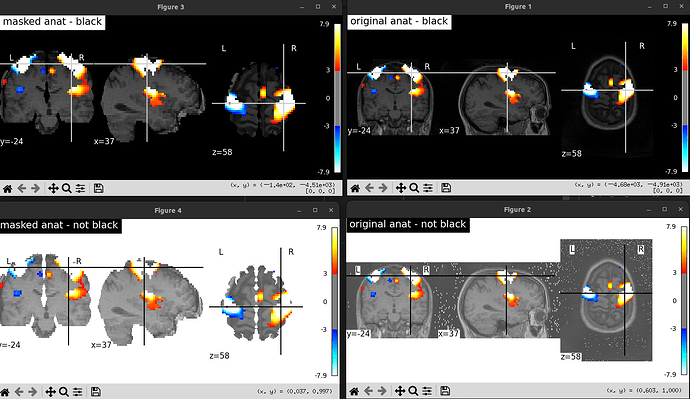
I wanted to plot statistical maps on MNI Pediatric template. I need a white background (black works as intended) however I get weird grey squares around the brain slices. As far as I know, the T1w image I have is fine, and it works if I use the default nilearn template. Passing the background image as path or a nifti object gives the same result.
import matplotlib.pyplot as plt
from nilearn.plotting import plot_stat_map, view_img
from nilearn.image import load_img
import os
from nilearn.glm import threshold_stats_img
from templateflow import api as tflow
import nibabel as nib
def plot_unc_figs(input_dir,output_dir):
contrast = ['Go correct>0','Stop correct>0','Stop error>0','Stop error>Go correct','Stop correct>Go correct']
grp = ['HC','OCD','OCD-HC']
bg_ = '/mnt/projects/TECTO/nihpd_asym_04.5-18.5_nifti/nihpd_asym_04.5-18.5_t1w.nii'
for i in contrast:
for g in grp:
zmap = load_img(os.path.join(input_dir,i,f'zmap_{i}_{g}.nii'))
thresholded_map, threshold = threshold_stats_img(
zmap,
alpha=0.001,
height_control="fpr",
cluster_threshold=10,
two_sided=True,
)
figure=plot_stat_map(thresholded_map,bg_img=bg_,draw_cross=False, black_bg=False, threshold=threshold)
figure.savefig(os.path.join(output_dir,f'{i}_{g}.tiff'),dpi=300)
display = view_img(thresholded_map,bg_img=bg_,draw_cross=False, black_bg=False, threshold=threshold)
display.save_as_html(os.path.join(output_dir,f'{i}_{g}.html'))
plt.close()
if __name__ == "__main__":
input_dir ='/mnt/scratch/projects/TECTO/bids_func/derivatives/fmriprep/SecondLevel_25/acompcor'
output_dir = '/mnt/projects/TECTO/SST_baseline_10_sep_2024/figures/fMRI/uncorrected'
os.makedirs(output_dir,exist_ok=True)
plot_unc_figs(input_dir,output_dir)
Thank you.
Best,
Vytautas