I’ve begun exploring and experimenting with (very) pilot data from a new 7T scanner. Nearly every run uses a slightly different acquisition protocol; combinations of 3-5 echoes, multiband acceleration of 4-6, in-plane GRAPPA acceleration of 2. (Apologies for the vagueness; I’m happy to share details/files/etc., but didn’t want this to be even longer.)
Getting towards my question: using fMRIPrep for preprocessing (command below), when I include multiple echoes in the dataset it writes a single bold.nii.gz as expected, with accompanying T2starmap.nii.gz (and many other files). Most of the outputs look fine, but the combined 4d bold and T2starmap have matching very obvious black lines:
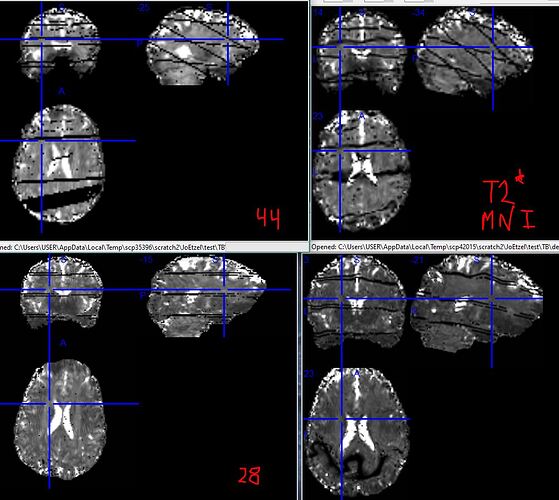
Above are examples for two runs (same person and session, different acquisitions), 44 (top row) and 28 (bottom row). The left column is subject space (/t2s_reporting_wf/clip_t2star/T2starmap_clipped.nii.gz), right MNI (_space-MNI152NLin2009cAsym_T2starmap.nii.gz). Both of these runs were multiband 4, and the horizontal lines in subject space are where the multiband boundaries ought to be. I don’t know offhand why 44 has diagonal lines and 28 does not, but the perfect geometry makes it seem safe to assume it has something to do with the acquisition parameters. (MB5 and MB6 runs have correspondingly more horizontal lines in their output.)
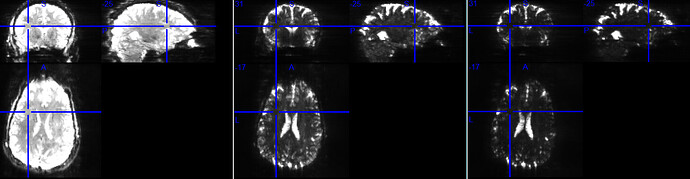
These black lines are only so obvious in the tedana-combined output, not if fmriprep is run with only one echo in the BIDS dataset, nor if its call includes --me-output-echos. Temporal standard deviation images (below) of the functional runs before preprocessing look more or less as I expected; these are for the three run 28 echoes (same color scaling in all three):
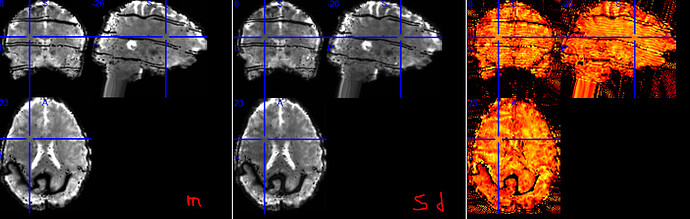
I suspect tedana is identifying the multiband boundaries (and maybe something GRAPPA-related?) as noise; their removal causing the black lines in the combined image. For a final example, below are temporal mean, standard deviation, and tSNR images of the combined run 28 (_run-28_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz):
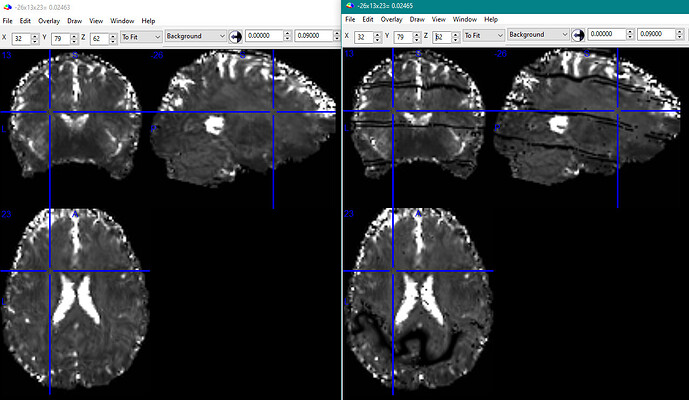
The actual questions (![]() ): Is this sort of “striping” behavior expected in optimally combined images (a sign of too much acceleration, perhaps?), or a bug somewhere in my logic, procedures, or the software? Any suggestions for how to investigate further?
): Is this sort of “striping” behavior expected in optimally combined images (a sign of too much acceleration, perhaps?), or a bug somewhere in my logic, procedures, or the software? Any suggestions for how to investigate further?
We have another pilot session scheduled next week, so can try different acquisition parameters; any suggestions? These stripes have appeared in all runs I’ve processed so far; three different participants and sessions.
Thanks!
fMRIPrep version: 25.1.0
fMRIPrep command: /opt/conda/envs/fmriprep/bin/fmriprep --fs-license-file /tmp/.license/freesurfer/license.txt -w /tmp /tmp/BIDS /tmp/derivatives participant --participant_label TB --output-spaces MNI152NLin2009cAsym --n-cpus 16 --omp-nthreads 4 --mem-mb 64000 -v --fs-no-reconall