Hi all,
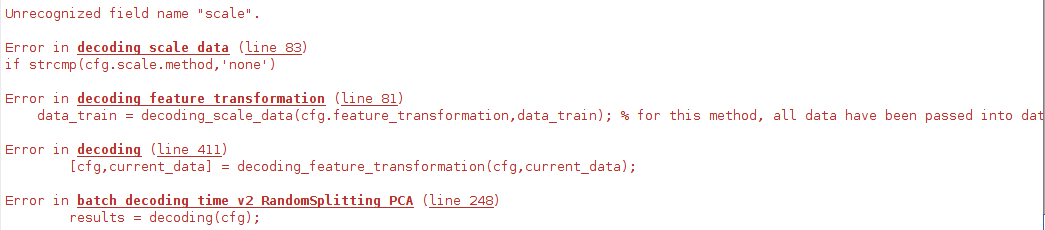
I use the TDT in matlab, and I dont know why it does not exist the file "accuracy_minus_chance.mat" when I add the 3 lines of PCA.
Here are the codes of configuration of TDT.
cfg = decoding_defaults;
cfg.analysis = 'ROI'; % 'wholebrain' but limited to the ROI using masks
cfg.software = 'SPM12';
%cfg.results.output = {'accuracy_minus_chance'}; % 'accuracy_minus_chance' by default
cfg.results.dir = outdir;
cfg.results.filestart = ['perm' sprintf('%04d',0)];
%cfg.results.write = 0; % Set to 0 to avoid saving output files by default
cfg.results.overwrite = 1; % overwrite allowed
%cfg.plot design = 0; % Cross-validation diagrams are neither generated nor saved
cfg.verbose = 0;
cfg.plot selected_voxels = 0; % 0: no plotting, 1: every step, 2: every second step, 100: every hundredth step...
cfg.files.name = dtFiles; % a 1xn array of file names of the selected subject
cfg.files. label= fileLabels; % a nx1 array
cfg.files.chunk = fileChunks; % a 80x1 vector of lables, for each subject
cfg.files.mask = fileMasks; % Select the correspondant roi masks
cfg.design = make_design_cv(cfg); % Automatic creation of the leave-one-run-out cross validation design
%% Random Splitting (RS) Design
% This creates a design where all combinations of ntrain trials are integrated in the design
% L00 ntrain = ntrial-1
% Matrices train and test of size = [ntrial x length(labels)] x [nchunk]
% Here [40 (trials: 20x2runs) x 2(conditions: sweet or water)] x 40 (chunks) = 80x40
nsplit = nchoosek(ntrial, ntrain); % All combinations of ntrain trials
% C = nchoosek(1:ntrial, ntrain); % All combinations of ntrain trials, array exceeds maximum array size preference in this case
if nsplit<nchunk
disp("Rechoose nchunk");
end
train_run = zeros(ntrial, nchunk); % Initialize the train matrix for each run
for i 1:nchunk
train_trials = randperm(ntrial, ntrain); % Trials used for training
train_run(train_trials, 1) = 1; % Set the corresponding entries to 1
end
train = [train_run; train_run];
test = -train; % All trials belong to train or test
% Affectation in the cfg.design structure
cfg.design.label= repmat(fileLabels, 1, nchunk);
cfg.design.set = repmat(cfg.design.set(:,1), 1, nchunk);
cfg.design.test = test;
cfg.design.train = train;
cfg.design.function.name = mfilename; % Put current script name
cfg.design.function.ver = date; % Put current date
And below the 3 lines of PCA I want to add
I don’t know exactly how to set the feature.transformation.critical_value, so I just give it to 0.
But it does not generate the “accuracy_minus_chance.mat” any more.
cfg.feature_transformation.method = 'PCA';
cfg.feature_transformation.estimation = 'all';
cfg.feature_transformation.critical_value = 0; % only keep components that explain at least 10 percent variance
And also, I would like to use the permutation after PCA, but it seems that after adding the PCA, none of the exchanges were performed.
I am confused BECAUSE everything was fine until the three lines of code about PCA were added. Thanks in advance for the responses.
SSS