Dear TDT users
I have a question whether the design was correctly made and how to perform permutations. I have 146 subjects with CBF (perfusion) maps during two different conditions and I want to classify the two different conditions with the TDT (the maps are normalized in MNI space and corrected for global CBF-values).
I started the script with the decoding_template_nobetas.m and decoding_template_between subjects.m scripts and defined the following.
path_to_group = ‘/server/fo2-22/data/CBF_all_data/CBF_post_minus_pre/TDT_class/data_dir’;
group1_files = spm_select(‘FPList’, path_to_group, ‘^.*pre_gm04_res3.nii$’); % Condition 1
group2_files = spm_select(‘FPList’, path_to_group, ‘^.*post_gm04_res3.nii$’); % Condition 2
data_files = [cellstr(group1_files); cellstr(group2_files)];
cfg = decoding_defaults();
cfg.analysis = ‘searchlight’
cfg.results.overwrite = 1;
cfg.verbose = 2;
cfg.classification.method = ‘libsvm’;
cfg.decoding.method = ‘classification_kernel’;
cfg.labelname = {‘Post’, ‘Pre’};
label_all=[ones(146,1);-ones(146,1)] %first 146 post then 146 pre
cfg.labels = label_all;
cfg.results_dir = output_dir;
cfg.scale.method = ‘min0max1’;
cfg.scale.estimation = ‘all’;
cfg.files.label=label_all;
cfg.scale.estimation=‘all’;
cfg.files.chunk=[(1:146) (1:146)]; % that means each subjects has their own chunck number
cfg.design.unbalanced_data =‘ok’; %unclear why script gave an error because 146 images in both cases
cfg.files.mask = ‘/server/fo2-22/data/CBF_all_data/CBF_post_minus_pre/TDT_class/masks/GM_thresh04_res3_bin.nii’;
cfg.files.name=data_files;
% using make_design_boot_cv compared to make_design_cv yielded total different results
%cfg.design = make_design_boot_cv(cfg,100,1) %
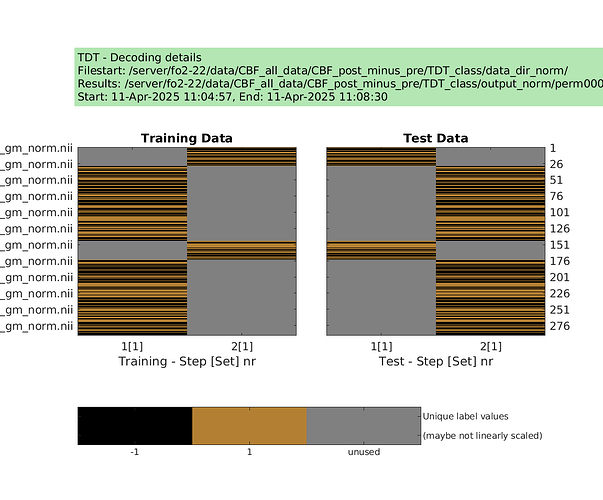
cfg.design = make_design_cv(cfg) % I decided to use this approach because with fix pairs (leave one subject (pair) out)
cfg.results.output = (‘AUC_minus_chance’,‘Accuracy_minus_chance’,'balanced_accuracy))
%cfg.results_dir = ‘/server/fo2-22/data/CBF_all_data/CBF_post_minus_pre/TDT_class/output_dir’;
cfg.results.dir = ‘/server/fo2-22/data/CBF_all_data/CBF_post_minus_pre/TDT_class/output_dir’;
display_design(cfg)
results = decoding(cfg)
Then I tried to run permutation test using the examples from make_design_permutation
cfg = rmfield(cfg,‘design’); % this is needed if you previously used cfg.
cfg.design.function.name = ‘make_design_cv’;
n_perms = 1000; % pick a reasonable number, the function might compute less if less are available
combine = 0;
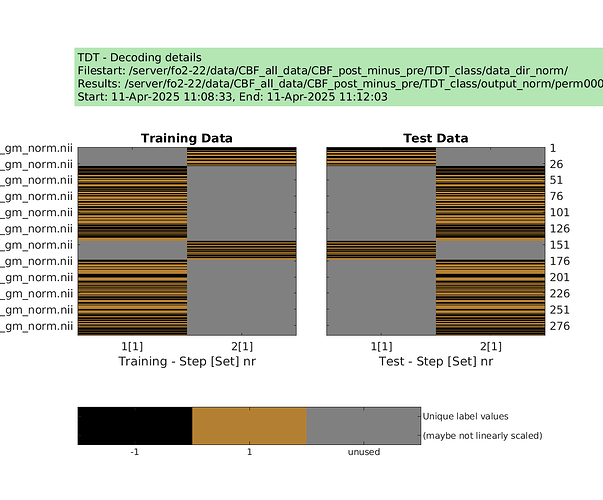
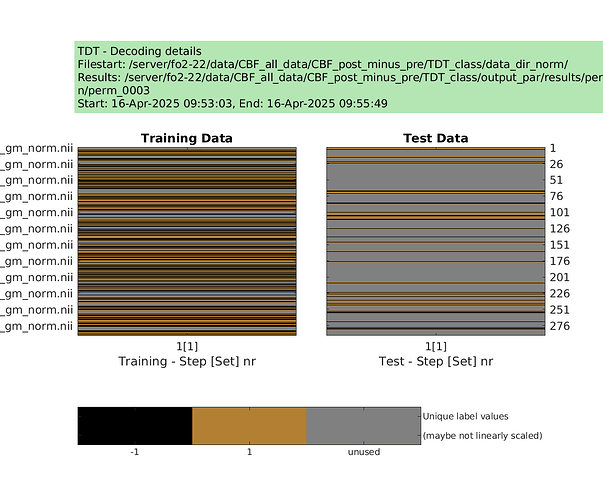
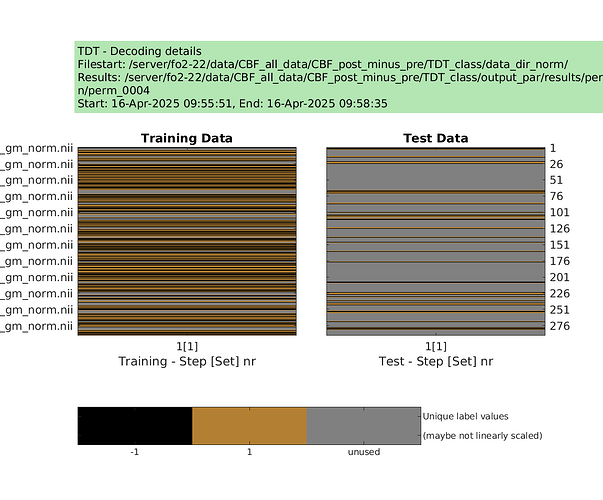
designs = make_design_permutation(cfg,n_perms,combine);
for i_perm = 1:n_perms
cfg.design = designs{i_perm};
cfg.results.dir = [‘/server/fo2-22/data/CBF_all_data/CBF_post_minus_pre/TDT_class/output_norm/perm’ sprintf(‘%04d’,i_perm)];
decoding(cfg); % run permutation
end
However, after one day only 6 permutations were finished. How can I adjust the script to speed up the process? It’s not clear for me how to run in parallel mode (parallel pool in matlab). Should I run several make_design_permutations and combine them later?
When every subject is an own chunk (like in this design), what is the best solution?
Moreover, I was wondering about negative values in res_AUC_minus_chance and res_accuracy_minus_chance maps (with values less than -25).
Best,
Ralf