Thanks for clarifying, things make a bit more sense but I unfortunately I have more concerns. Lets take it from the top though.
For your short run - denoising is likely not appropriate. You can get a tSNR from the ‘optimal combination’ (Using tedana from the command line — tedana 0.0.12+0.g863304b.dirty documentation) without denoising. I’m not completely sure what your goal is when you say a “test on the flip_angle” but if you are varying the flip angle, getting 35 volumes and then repeating that process - then you would want to see tSNR without denoising anyway. That way it is a fair comparison.
For tedana results - I’m very worried about your preprocessing. If 3dunifize is being used on each echo independently that is probably wrong - it is altering the very important echo-specific information. If you are just applying it to a dataset to create a mask, then not using the unifized data, that would be ok.
Even then, it is also not clear why you are applying a brain mask as an early step. That seems like it is unnecessary.
Are you estimating the parameters from one echo with MCFLIRT them applying that to all of them? Motion correction should not be ran on each echo independently.
Despike should probably be a first step, rather than the last step before tedana.
With a 4 seconds TR, you should be using slice timing correction.
Your normalized data being too big for tedana is likely because when you apply the ants transforms you are writing out super small voxels, which is just making your datasets huge with no purpose (upsampling during processing can be useful in very high resolution scenarios, but not here).
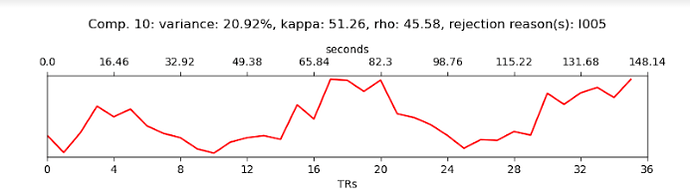
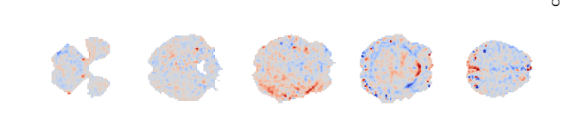
For the second dataset I am even more concerned. You have enough timepoints, but I see very little meaningful structure there, other than the artifact previously noted. Given all of the potential issues above, it is may not be very useful to look at the tedana output.
The only thing I can say there is that it looks like the MRI sequence is potentially not good. You have a very long TR (despite using multiband??), but also strange artifacts that show up in many of the components. The brain mask that you have used looks like it may also not be very good - but again, it is kind of hard to tell from just the components.
What are your MR sequence parameters (GRAPPA, multiband, voxel size, field of view, bandwidth, etc etc)? It looks almost like you are using multiband 4, but to do that and still have a 4s TR …well, that is extraordinarily confusing. What does each echo look like? How many channels does your head coil have?
tedana will work on datasets of varying size, but 35 volumes could be too short for denoising. The small datasets may be too small, but they may also just be bad data.
I would recommend using afni_proc.py AFNI program: afni_proc.py - consider example 12 and its various approaches. It looks like you already have afni installed. You could also use fmriprep but I don’t know much about that.
Assuming that the MRI data itself can be salvaged…I would suggest processing the data in a very very simple way first, like the following.
Slicetime correct each echo
Motion correct the first echo and apply those parameters to echoes 1,2,3,4
tedana
You can then take 3dAllineate, ANTS etc - those steps seem fine except for the possibility that you are taking a 3x3x3mm dataset to 1x1x1. I believe there is an option for a reference volume when applying ants transforms.
I would take the MNI template and resample it to your fMRI voxel size (3dresample -dxyz 3 3 3 -prefix resampled_MNI_template.nii.gz -input Your_MNI_template_image.nii.gz ) and use that as a reference image, assuming your voxels are indeed 3x3x3 in the example provided.
![]()