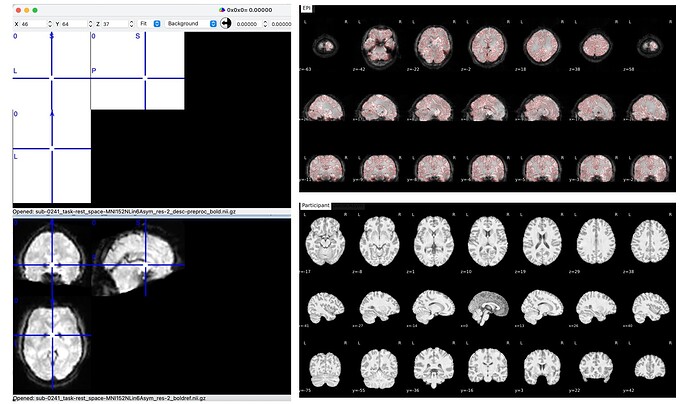
Summary of what happened:
Hi everyone!
I am trying to preprocess resting state fMRI data using fmriprep (Singularity on HPC, v24.0.1). All data runs fine without errors. However, when I visualized it, I found that the spatially normalized data for some subjects looked abnormal. The data with the suffix *res-2_desc-preproc_bold.nii.gz is very small (< 1mb). I cannot see the normalized functional images when opening it with MRIcron.
I tried to inspect the html about the registration, but it looked normal.
I would like to know what causes this situation, and thank you all for your time and effort on this!
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity exec --cleanenv --bind '$'{templateflow}:/data/templateflow --bind '$'{input_dir}:/data/input --bind '$'{output_dir}:/data/output '$'{singularity_image} fmriprep --participant-label ${sbj} --output-spaces MNI152NLin6Asym:res-2 MNI152NLin2009cAsym:res-2 --nthreads 2 --stop-on-first-crash --notrack -w /path/to/working/ --bids-database-dir '$'{input_dir} --fs-license-file '$'{fs_license} --fs-no-reconall --skip_bids_validation '$'{input_dir} '$'{output_dir} participant
Version:
24.0.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE