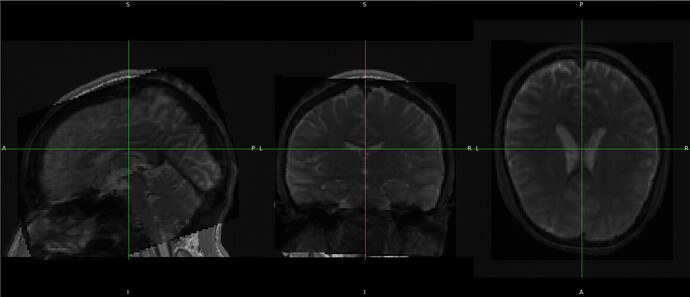
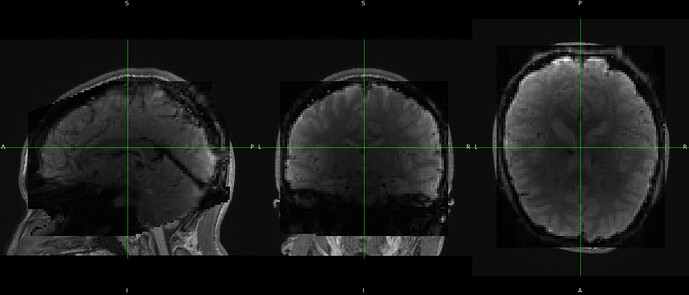
Hi, @Steven
Thank you for your prompt response! May I bother you with a few more questions?
1. Modification of the Input T1w File for fmriprep
We are currently utilizing both qsiprep and fmriprep to process our multi modal data. However, we encountered a disparity in space between the results obtained from qsiprep (in T1w ACPC space, a 6DOF transformation and resampling) and fmriprep (in T1w Native space). Our goal is to achieve spatial alignment between the outputs of fmriprep and qsiprep.
Here is our proposed solution: we use the results from qsiprep to register the original T1 file to the ACPC space, denoted as T1w_native_acpc. Subsequently, we overwrite the original T1w file in the BIDS with T1w_native_acpc and execute fmriprep based on this modified bids.
We have observed that fmriprep runs successfully, and the preprocessed images (T1w and Bold) are aligned with the qsiprep ACPC space. We have conducted some tests to compare our approach with fmriprep’s original results (denoted as raw) in standard spaces (MNI2009cAsymLin, fsLR) for voxel/vertex to voxel/vertex comparison. We found a high similarity in volume results, while there are some discrepancies in the results under fsLR surface space. However, the overall patterns (functional connectivity, default network) remain consistent. Here are some examples.
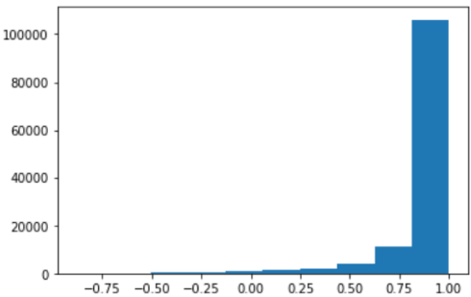
The Pearson correlation coefficient distributions of voxel time series between raw and our method in MNI
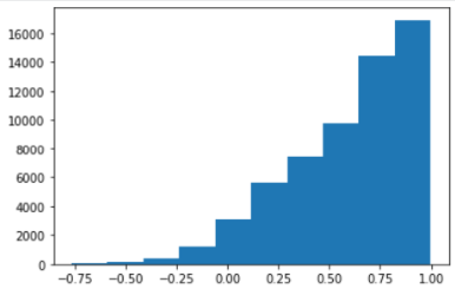
The Pearson correlation coefficient distributions of vertex time series between raw and our method in fsLR seems unsatisfactory.
So, we tested the overall pattern.Here is the functional connectivity matrices from the raw and our method in fsLR(left as our mehod, right as raw)
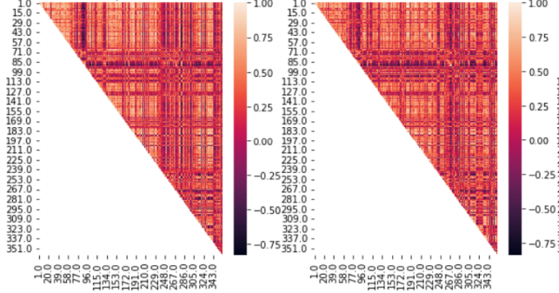
the correlation between FC matrix is 0.846
We would appreciate your insights on whether you consider this method/solution feasible and if you have any other concerns regarding this strategy.
2. Out of curiosity, you mentioned that fmriprep does not use T1w JSON information. Could you clarify if it utilizes JSON information from the BOLD files?
Please let me know if there is anything unclear!Thank you for your time!
Best regards,
Jingli