Hello,
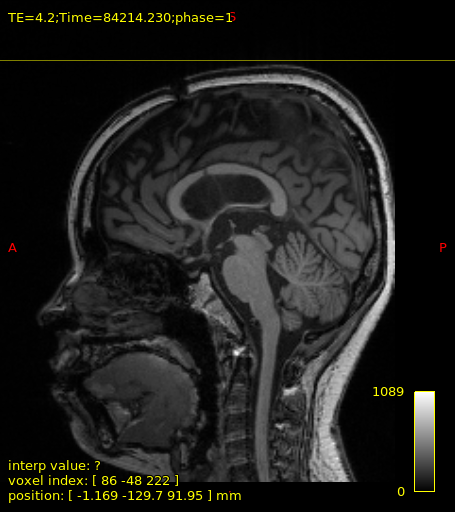
Within my Dataset, a number of Patients have severely displaced or discontinuous white matter tissue.
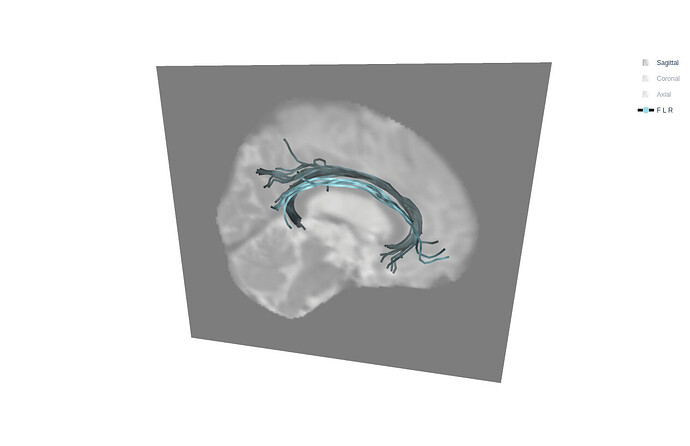
After analyzing the T1 images, this seems to factor impairing my tractography targeting the fornix using pyAFQ recobundles.
From my current knowledge, using a NOT ROI would get rid of CGC fibers, which are the most prominent source of erroneous fibers, sometimes leading to the whole CGC being extracted instead of the fornix like intended(or at least together with fornix). Since recobundles uses a Streamline atlas as reference, it does not seem to deal well with displaced fibers. This leads me to the following questions:
- Are there any tools to increase the accuracy of the recobundles within pyAFQ specifically regarding this scenario? Especially for discontinuous tracts.
- Additionally, is there a way to remove a low number(3-8) fibers, which are from a different tract, post tractography?
- Sometimes it even grabs the CGC(exclusively or together with fornix) with regular tissue locations, I do not have an explanation for those cases, any ideas?
Here is an example T1 image to highlight the situation a bit:
Thanks and cheers,
Christian