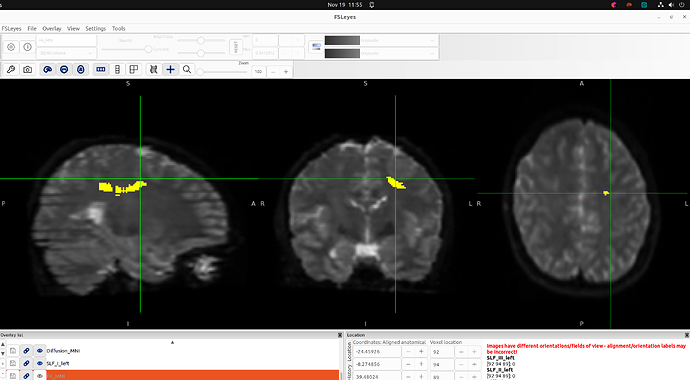
Summary of what happened:
I’m trying to use TractSeg (GitHub - MIC-DKFZ/TractSeg: Automatic White Matter Bundle Segmentation) to segment white matter tracts from diffusion data that was preprocessed with QSIprep.
Initially, I wasn’t sure whether I needed to convert my data to MNI space, since the TractSeg documentation states:
“For best results the input image must have the same orientation as the HCP data (MNI space)… If the image orientation and the gradient orientation of your data is the same as in
examples/Diffusion.nii.gzyou are fine.”
My diffusion image has the same orientation as the example file, so I assumed a transformation to MNI space wasn’t necessary. However, I tested both approaches:
- Running TractSeg with the MNI transformations
- Running it without any MNI transformation
In both cases, my commands ran without errors, but when generating bundles with --nr_fibers, TractSeg reported that the tract masks were empty, and thus produced empty tractograms, even though the masks are visible when I open them in FSLeyes (see image).
I’m wondering if this issue is related to the space of the QSIprep outputs, or possibly to my bval/bvec transformations not being applied correctly. I’m not fully sure where the pipeline is breaking.
Any advice or suggestions would be greatly appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
#Transformation to MNI space for TractSeg
calc_FA -i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.nii.gz -o .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/FA.nii.gz --bvals .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.bvals --bvecs .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.bvecs \
--brain_mask .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/nodif_brain_mask.nii.gz
flirt -ref .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/MNI_FA_template.nii.gz -in .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/FA.nii.gz \
-out .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/FA_MNI.nii.gz -omat FA_2_MNI.mat -dof 6 -cost mutualinfo -searchcost mutualinfo
flirt \
-ref .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/MNI_FA_template.nii.gz \
-in .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.nii.gz \
-out .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/Diffusion_MNI.nii.gz \
-applyxfm -init FA_2_MNI.mat \
-dof 6
cp \
.../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.bvals \
.../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/Diffusion_MNI.bvals
rotate_bvecs \
-i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/sub-02_acq-b0b1000b2000_dir-AP_space-T1w_desc-preproc_dwi.bvecs \
-t FA_2_MNI.mat \
-o ...BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output/Diffusion_MNI.bvecs
# Commands to run TractSeg
TractSeg -i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/Diffusion_MNI.nii.gz -o .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/ --raw_diffusion_input --output_type tract_segmentation
TractSeg -i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/peaks.nii.gz -o .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/ --output_type endings_segmentation
TractSeg -i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/peaks.nii.gz -o .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/ --output_type TOM
Tracking -i .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/peaks.nii.gz -o .../BIDS/derivatives/qsiprep_dering/qsiprep/sub-02/dwi/tractseg_output_MNI/tractseg_output/tractseg_output/ --nr_fibers 5000
Environment (Docker, Singularity / Apptainer, custom installation):
Docker from GitHub:
sudo docker run -v /absolute/path/to/my/data/directory:/data \
-t wasserth/tractseg_container:master TractSeg -i /data/my_diffusion_file.nii.gz -o /data --raw_diffusion_input