Summary of what happened:
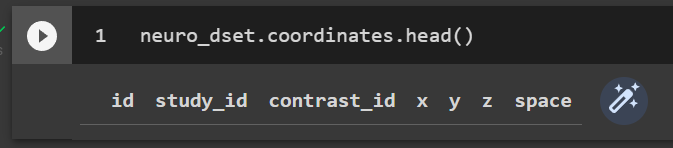
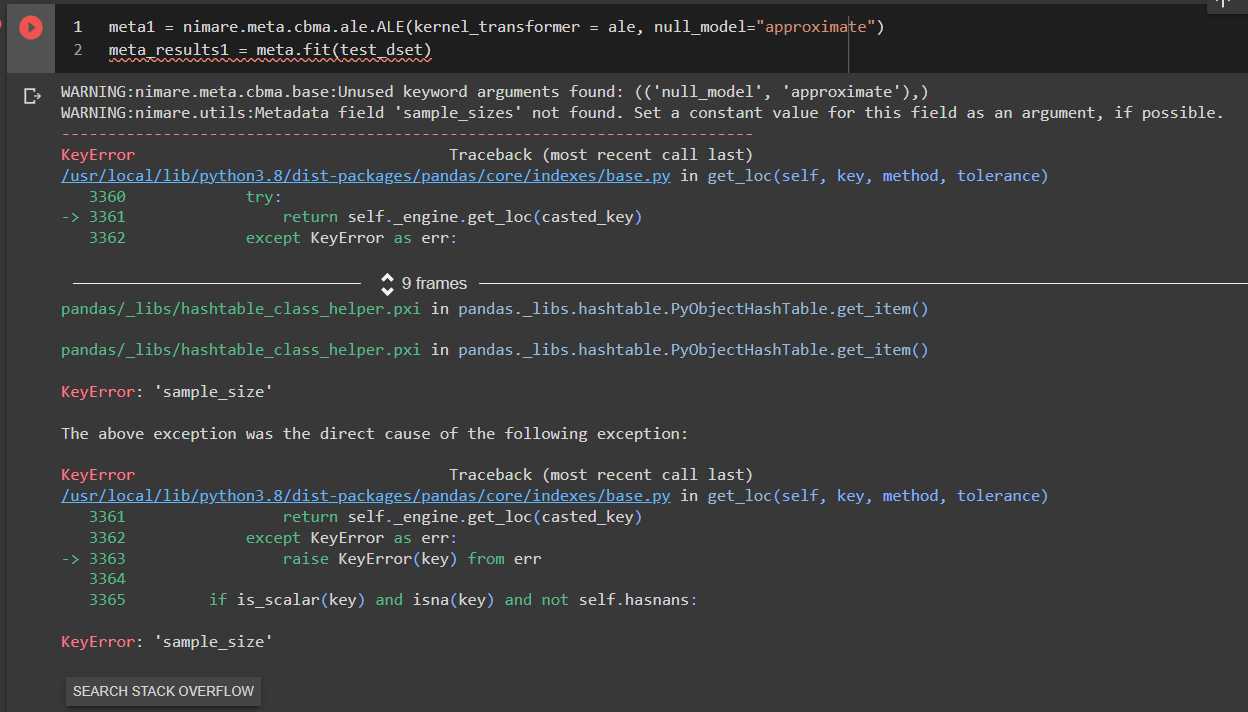
I am struggling to get the coordinates from four studies I want to analyze from Neurosynth. I downloaded the full Neurosynth database and then used the slice() method with ids, stored in a 1D numpy array. Even though when I print the new data set it says it includes four experiments, when I try to view their ids, coordinates, etc. the sections are empty.
Command used (and if a helper script was used, a link to the helper script or the command generated):
import numpy as np
ids = np.array([21568643,26610651,11087002,18510438])
neuro_dset = neurosynth_dset.slice(ids)
neuro_dset.save(os.path.join(out_dir, "sliced_neurosynth.pkl.gz"))
neuro_dset.coordinates.head()
Version:
The version was python 3.8.10
Environment (Docker, Singularity, custom installation):
I did everything in Google Colab because for some reason I could not install nimare on my machine.
Data formatted according to a validatable standard? Please provide the output of the validator:
I used NiMARE’s documentation’s recommended way of extracting the data from Neurosynth and converting it into a .pkl.gz dataset:
out_dir = os.path.abspath("/example_data/")
os.makedirs(out_dir, exist_ok=True)
files = fetch_neurosynth(
data_dir=out_dir,
version="7",
overwrite=False,
source="abstract",
vocab="terms",
)
# Note that the files are saved to a new folder within "out_dir" named "neurosynth".
pprint(files)
neurosynth_db = files[0]
neurosynth_dset = convert_neurosynth_to_dataset(
coordinates_file=neurosynth_db["coordinates"],
metadata_file=neurosynth_db["metadata"],
annotations_files=neurosynth_db["features"],
)
neurosynth_dset.save(os.path.join(out_dir, "neurosynth_dataset.pkl.gz"))
Relevant log outputs (up to 20 lines):
“Dataset(4 experiments, space=‘mni152_2mm’)”
Screenshots / relevant information: