Summary of what happened:
I have been performing quality checks on data I preprocessed with TrUE-NET’s prepare_truenet_data command, and I am noticing a number of issues. All preprocessing steps are performed in FSL.
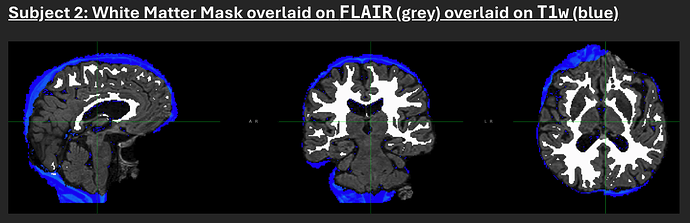
First, skull-stripping is poor and inconsistent between FLAIR and T1w modalities. In general, FLAIR skull-stripping is too aggressive and T1w skull-stripping is too liberal. In both cases, a lot of neck is left behind, though different parameters seem to have been used in skull-stripping each image, resulting in images of different sizes and tissue coverage. Since the brain mask is based on the skull stripped T1w, it generally covers a lot of non-brain tissue.
Second, the white matter mask generated by FAST/BIANCA seems sufficient for capturing deep white matter around the ventricles but is less thorough in covering white matter more peripherally through the gyri. I know white matter hyper-intensities tend to be periventricular, so I’m not sure how much of a problem this actually is, but it does not seem ideal.
The T1w registration to FLAIR space and bias correction all look good.
Are there options built into prepare_truenet_data that I could use to resolve these issues, or is there an alternative method for image preprocessing that I could use on the images before running TrUE-NET? Since I don’t fully understand how TrUE-NET works, I’m not sure to what extent these issues represent critical failures in preprocessing.
Command used:
prepare_truenet_data
Version:
1. FSL version 6.0
2. TrUE-NET GitHub: