Hello,
I am trying to wrap my head around q-form and s-form matrices, and while I have theoretical knowledge about the same, I need help interpreting the values I see on the header files. Can you please help me understand what these values mean and if I should be worried about the differences I see?
Procedure – the images were acquired with a Siemens 3T scanner using syngo MR XA30 software with phase encoding in the PA direction. The Dicoms were converted to Nifti using the dcm2niix function and renamed to fit the HCP naming and folder convention.
Approach: Preprocess my data using HCP pipeline > Convert preprocessed EPI to native space using applywarp > Do 1st-level analysis > Convert the statistical dataset back to MNI to extract ROI-specific beta series and compute the correlation.
Within my unprocessed data
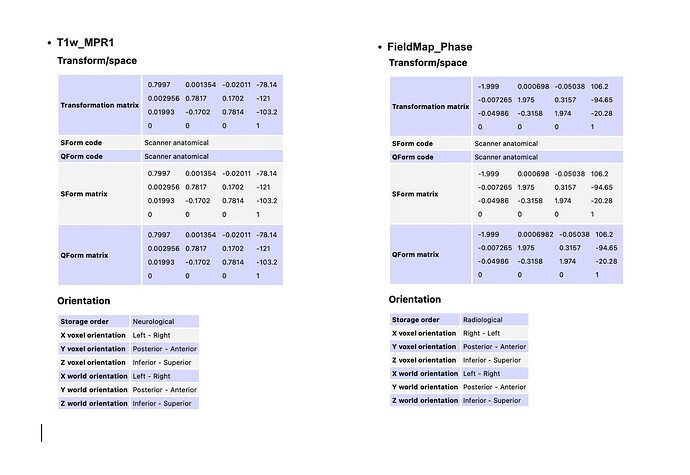
- My T1w_MPR1 and my field maps have different x voxel orientations, with T1w_MPR1 having a left-right orientation and the field map phase and magnitude images having a right-left orientation. Does x_voxel orientation indicate the order in which values are stored in the file or that the x coordinates are positive on the left side?
-
My unprocessed EPIs are also in the right-left x voxel orientation with storage order indicating radiological. Is this difference in x-voxel orientation between the unprocessed T1w and EPI normal?
-
After the HCP pipeline, my T1w and T2w within the MNINonLienar folder have a right-left orientation for x_voxel orientation. Should this not be left-right since the MNI convention is LPI? Or am I understanding these values incorrectly?
Why is this important to me? I am trying to define ROIs using the Power 2011 atlas, a set of 264 coordinates in MNI. I need to understand the orientation correctly to ensure I am mapping the coordinates to the correct side of the brain.
Can you please help me understand this topic better? Thank you for taking the time to review my questions.