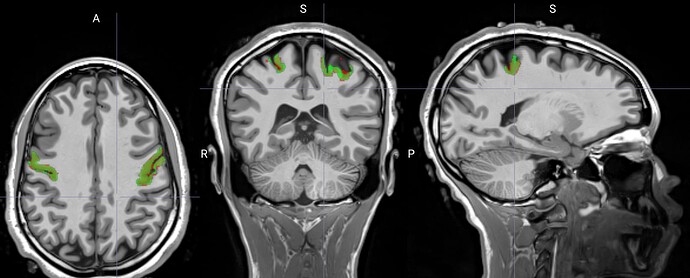
Hello,
I was wondering whether I am correctly using the nilearn.image.resample_img function. I want to extract BOLD timeseries from within a particular mask (ROI) using the code below:
data_masked_scan2_localizer = [masking.apply_mask(i, vis_mask, smoothing_fwhm=csmooth) for i in data_task_scan_loc2]
where i is the list of functional BOLD images (fmriprep: f'{mri_sub}*task-{mri_task_name}*T1w_desc-preproc_bold.nii) for which this needs to be performed and vis_mask the ROI mask.
The mask has been registered with the subjects T1w image using ANTSTransforms (this was done for the purpose of another set of analyses). This is the code I used for this:
os.system("antsApplyTransforms --default-value 0 --float --interpolation NearestNeighbor -d 3 -e 0 -n GenericLabel \
--input %s/%s.nii \
--reference-image %s/ANAT_T1w_images/%s/long_T1.nii \
--output %s/%s.nii \
--transform %s/y_MNI_to_T1w.h5 -v" \
Before extracting signal from my functional images I check whether the Affine align transformations are the same between mask and functional images. If not, I resample the mask using the following code:
mask_img_resampled = image.resample_img(vis_mask, target_affine=tmp_ref.affine, target_shape=tmp_ref.shape[:3],interpolation="nearest")
Is it correct that since both the mask and the BOLD functional images are already in the same space (T1w space), the only thing I need to do before extracting the signal from my functional images is resample the mask to the BOLD grid?
Nina