Summary:
2024-07-25T16:00:00Z
Hi all friends:
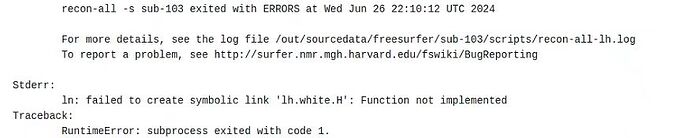
Recently I am using the latest version of fmriprep-docker to complete preprocessing of task-related fMRI data with the codes that worked on fmriprep-docker 23.2.1. However, two important steps (--use-aroma and --use-syn-sdc) went wrong and errors occurred. ![]()
after checking the documentations and posts in forum, I already know that --use-aroma had been marked [ DEPRECATED] Usage Notes — fmriprep version documentation and removed from the pipeline (although I stick to the fact that it is essential or at least an option for fMRI preprocessing!), but I have no idea why the latest docker version fmriprep cannot recognize the --use-syn-sdc command, at least 23.2.1 this command works normally and no documentation said that it was banned in 24.0. My question is similar to this post Use-syn-sdc not working for fmriprep 22.0.2 but not quite the same.
Here are my codes run on 24.0.1 and 23.2.1:
#24.0.1, aroma and SDC not working
#!/bin/bash
bids_root_dir=/media/weissley/HIKSEMI/MST
sublist=/media/weissley/HIKSEMI/listnum.txt
for sub in `cat $sublist`
do
echo -e "\033[32m---running sub-${sub} fmriprep--- \033[0m"
docker run --security-opt seccomp=unconfined --rm -it \
-v $bids_root_dir:/data:ro \
-v $bids_root_dir/derivatives:/out \
-v $bids_root_dir/codes/license.txt:/opt/freesurfer/license.txt \
-v $bids_root_dir/derivatives/work:/work \
nipreps/fmriprep:latest \
/data /out \
participant \
--participant-label ${sub} \
#--use-aroma \
#--slice-time-ref 0 \
--use-syn-sdc \
--fs-license-file /data/derivatives/license.txt \
--fs-no-reconall \
--output-spaces MNI152NLin2009cAsym:res-2 T1w\
--dummy-scans 0 \
-nifti \
--stop-on-first-crash \
-w /work
echo "all finished"
end
#23.2.1, aroma and SDC both works normally
docker run --security-opt seccomp=unconfined \
-v /expdata/jxh/data_LJ/4MI/BIDS/TG/:data \
-v /expdata/jxh/data_LJ/results/fmriprep/4MI/TG/:/out \
-v /expdata/jxh/data_LJ/results/fmriprep/4MI/TG/:/work \
-v /expdata/jxh/data_LJ/:/fs_licens \
nipreps/fmriprep:23.2.1 \
/data /out \
-w /work \
participant \
--participant-label 101 \
--use-aroma \
--use-syn-sdc \
--cifti-output \
--output-spaces MNI152NLin6Asym:res-2 T1w \
--fs-license-file /fs_licens/license.txt \
Version: 24.0.1
Environment: Docker, installed by pull images.
Data formatted
MST
Summary
* 204 Files, 2.76GB
* 10 - Subjects
* 1 - Session
Available Tasks
* MST
Available Modalities
* MRI
This is a valid BIDS dataset!
relevant information:
This is the information of error (apparently, the pipeline skipped the use-syn-sdc command and successfully finished):
240726-02:56:15,809 nipype.workflow IMPORTANT:
fMRIPrep finished successfully!
240726-02:56:15,811 nipype.workflow IMPORTANT:
Works derived from this fMRIPrep execution should include the boilerplate text found in <OUTPUT_PATH>/logs/CITATION.md.
Sentry is attempting to send 1 pending events
Waiting up to 2 seconds
Press Ctrl-C to quit
fmriprep.sh: 行 19: --use-syn-sdc: 未找到命令
all finished
Translation of Chinese: the line 19 of fmriprep.sh --use-syn-sdc command was not found or recognized.
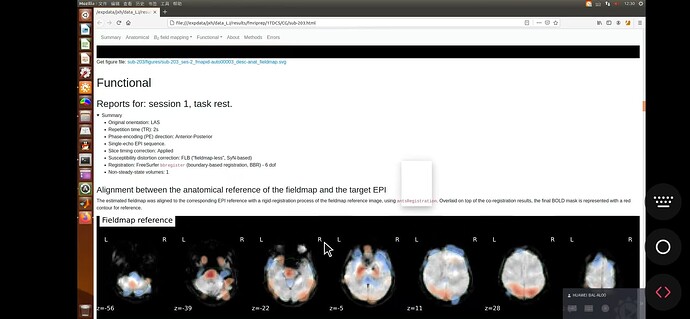
Here, the report also shows that no SDC was performed.
However, .sh on 23.2.1 finished with no error, and the figure was successfully generated——as follow. I would appreciate a lot if anyone can tell the reason!