Summary of what happened:
Hey all,
I’m using the mri_watershed algorithm to create a BEM model for MEG analysis.(mne.bem.make_watershed_bem — MNE 1.10.2 documentation)
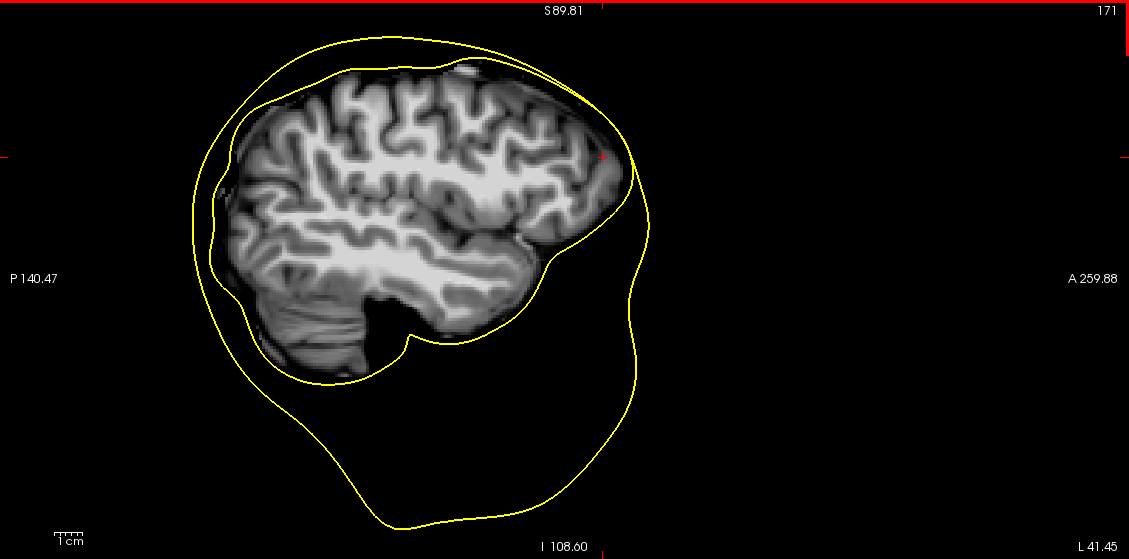
For a lot of subject, the process fails because the brain extraction step of the mri_watershed algorithm does not properly segment the brain and often keeps part of the dura matter. As a results, the expanded brain surface (i.e inner skull) is often not completely inside the outer skull area.
I’m using smrirpep to process my T1 images and the pipeline also outputs are more accurate brainmask ( refined using the aseg.mgz file: Processing pipeline details — fmriprep version documentation)
Is there a way to provide this brainmask to the mri_watershed command or is there another command that could reconstruct a brain surface from a brainmask ?
This idea would be to include this step in an automated nipype workflow.
Command used (and if a helper script was used, a link to the helper script or the command generated):
mri_watershed -h 10 -atlas -useSRAS -surf /data/proposal/derivatives/meegpype/sourcedata/freesurfer/sub-ENG008/bem/watershed/sub-ENG008 /data/proposal/derivatives/meegpype/sourcedata/freesurfer/sub-ENG008/mri/T1.mgz /data/proposal/derivatives/meegpype/sourcedata/freesurfer/sub-ENG008/bem/watershed/test.mgz
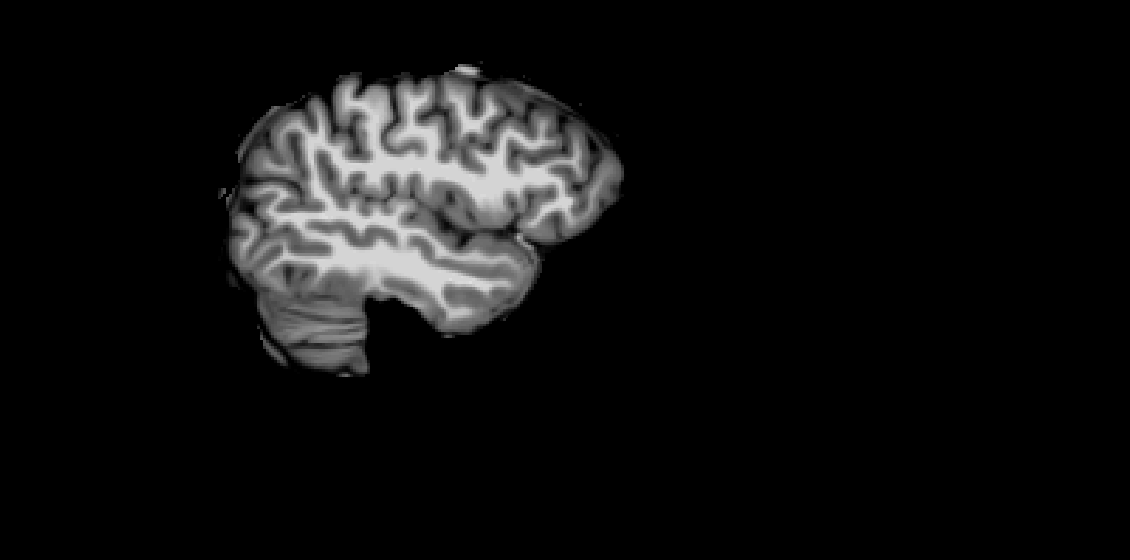
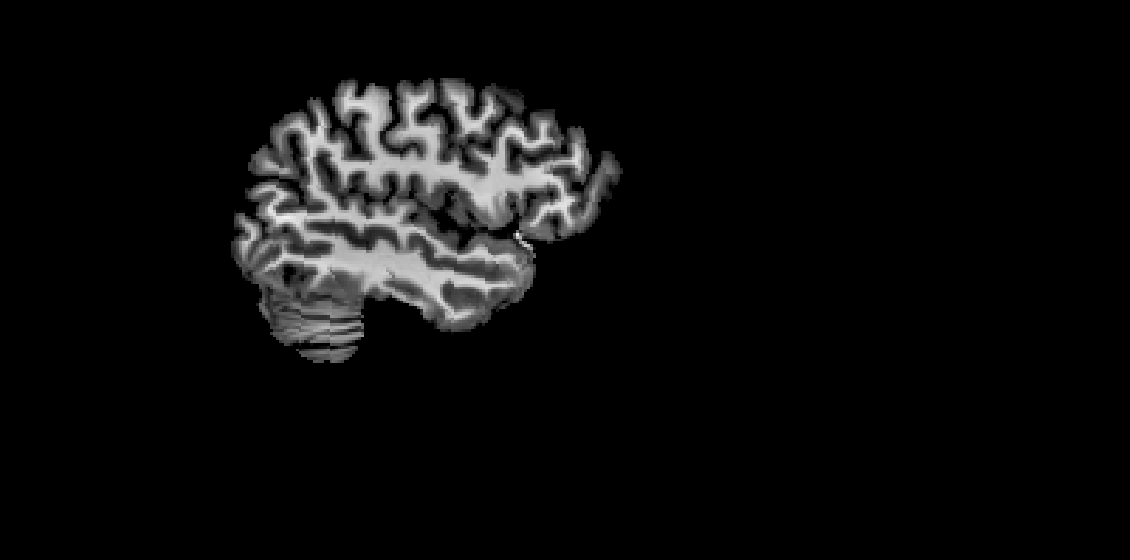
Screenshots / relevant information:
mri_watershed brain mask
smriprep brain mask
Thanks a lot for your help !