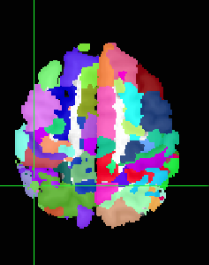
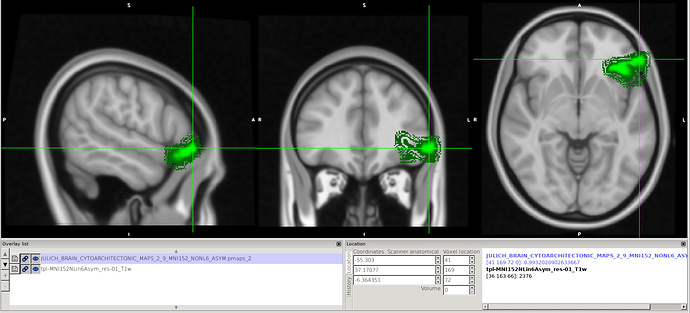
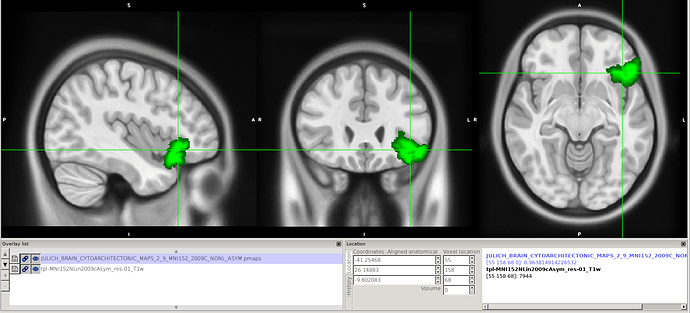
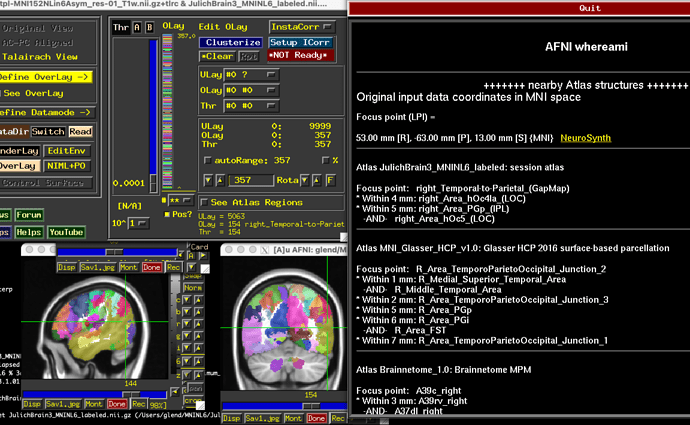
Hi @jsein @dglen and valeria! (sorry, new user, can’t mention more than 2 users)
We are trying to do the same thing but with a different atlas: MMP 1.0 MNI projections (MMP 1.0 MNI projections)
Indeed we have several atlases that we will need to register from 2009c to MNI152NLin6Asym.
We’ve tried running @jsein command using Ants 2.5.0, but it does not run. There are a couple of duplicated arguments (smoothing-sigmas, shrink-factors, etc).
In the end, I removed the second occurence of each and ran this:
antsRegistration \
- v 1 \
--collapse-output-transforms 1 \
--dimensionality 3 \
--float 1 \
--initialize-transforms-per-stage 0 \
--interpolation LanczosWindowedSinc \
--output '[ ants_2009cAsym_to_NLin6, ants_2009cAsym_to_NLin6_Warped.nii.gz ]' \
--transform 'SyN[ 0.1, 3.0, 0.0 ]' \
--convergence [ 100x70x50x20, 1e-06, 10 ] \
--smoothing-sigmas 3.0x2.0x1.0x0.0vox \
--shrink-factors 8x4x2x1 \
--use-histogram-matching 1 \
--winsorize-image-intensities '[ 0.005, 0.995 ]' \
--write-composite-transform 1 \
--metric 'Mattes[ tpl-MNI152NLin6Asym_res-01_T1w.nii.gz, tpl-MNI152NLin2009cAsym_res-01_T1w.nii.gz, 1, 56, Regular, 0.25 ]' \
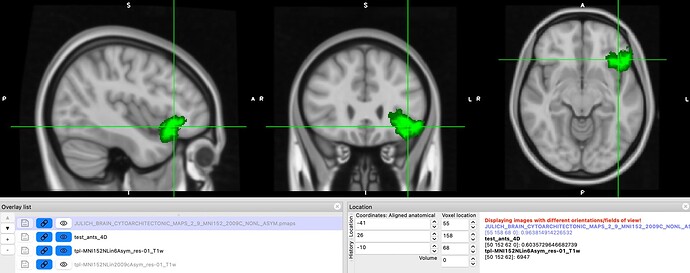
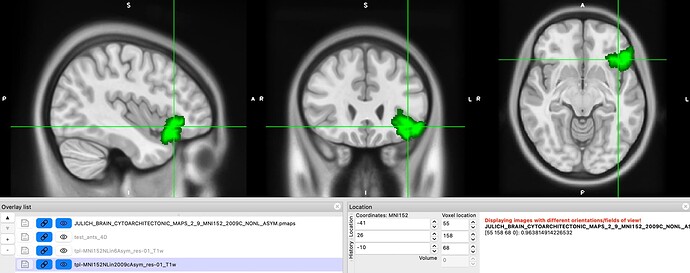
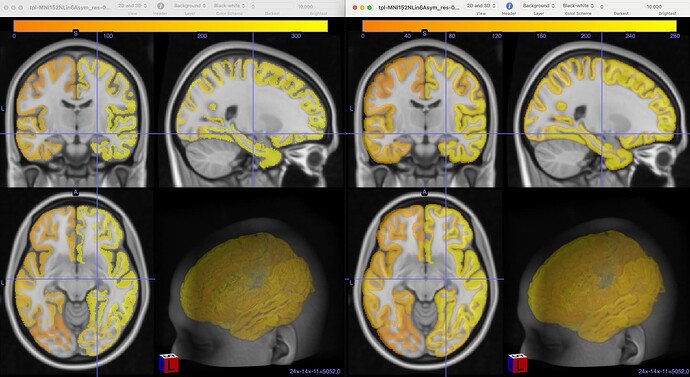
However, we still find some issues with the atlas after we applied the transform. In this image you can see on the left, the MNI152NLin6Asym template and atlas (warped). On the right, the MNI152NLin2009cAsym template and atlas (original)
The concerns comes mostly because we are “loosing” voxels, in some ROIs, more than 30% of them.
Is there any parametrisation that we can do to improve the accuracy of the transform?
FYI: we are still running @dglen approach, I’ll update as soon as it’s done.