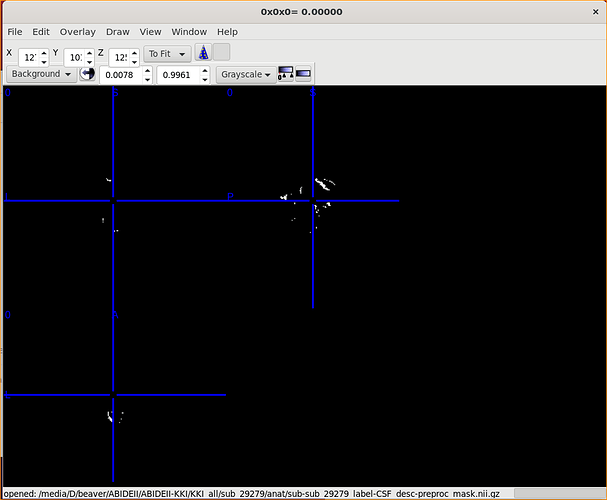
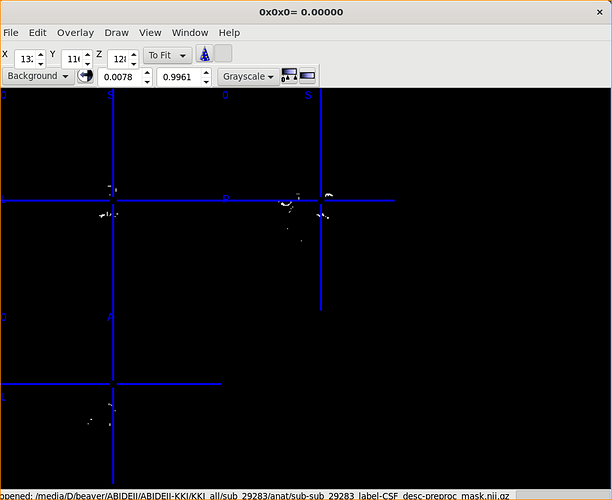
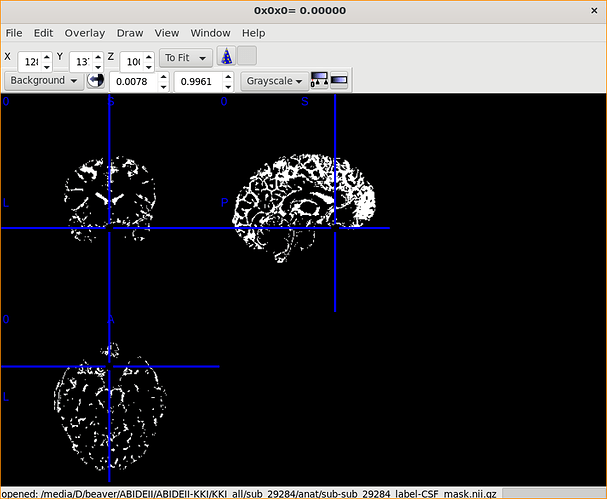
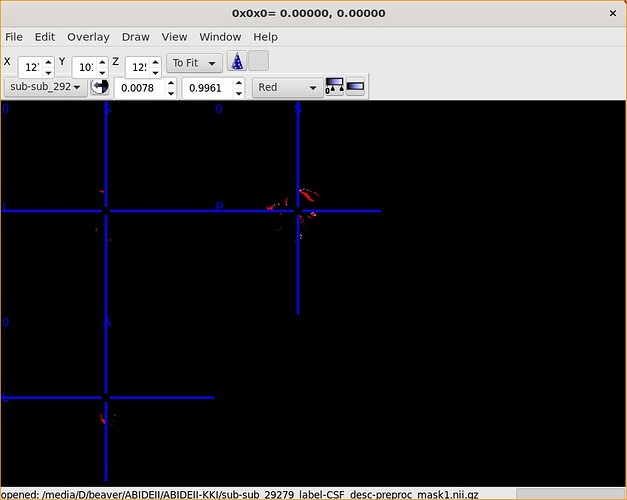
Of course!
here is my pipeline config file.
since I cannot upload attached files (new user), I am going to write it in code blocks.
%YAML 1.1
---
# CPAC Pipeline Configuration YAML file
# Version 1.8.1
#
# http://fcp-indi.github.io for more info.
#
# Tip: This file can be edited manually with a text editor for quick modifications.
pipeline_setup:
# Name for this pipeline configuration - useful for identification.
pipeline_name: cpac-default-pipeline
output_directory:
# Directory where C-PAC should write out processed data, logs, and crash reports.
# - If running in a container (Singularity/Docker), you can simply set this to an arbitrary
# name like '/output', and then map (-B/-v) your desired output directory to that label.
# - If running outside a container, this should be a full path to a directory.
path: /
# (Optional) Path to a BIDS-Derivatives directory that already has outputs.
# - This option is intended to ingress already-existing resources from an output
# directory without writing new outputs back into the same directory.
# - If provided, C-PAC will ingress the already-computed outputs from this directory and
# continue the pipeline from where they leave off.
# - If left as 'None', C-PAC will ingress any already-computed outputs from the
# output directory you provide above in 'path' instead, the default behavior.
source_outputs_dir: None
# Set to True to make C-PAC ingress the outputs from the primary output directory if they
# exist, even if a source_outputs_dir is provided
# - Setting to False will pull from source_outputs_dir every time, over-writing any
# calculated outputs in the main output directory
# - C-PAC will still pull from source_outputs_dir if the main output directory is
# empty, however
pull_source_once: True
# Include extra versions and intermediate steps of functional preprocessing in the output directory.
write_func_outputs: False
# Include extra outputs in the output directory that may be of interest when more information is needed.
write_debugging_outputs: False
# Output directory format and structure.
# Options: default, ndmg
output_tree: "default"
# Generate quality control pages containing preprocessing and derivative outputs.
generate_quality_control_images: True
working_directory:
# Directory where C-PAC should store temporary and intermediate files.
# - This directory must be saved if you wish to re-run your pipeline from where you left off (if not completed).
# - NOTE: As it stores all intermediate files, this directory can grow to become very
# large, especially for data with a large amount of TRs.
# - If running in a container (Singularity/Docker), you can simply set this to an arbitrary
# name like '/work', and then map (-B/-v) your desired output directory to that label.
# - If running outside a container, this should be a full path to a directory.
# - This can be written to '/tmp' if you do not intend to save your working directory.
path: /tmp
# Deletes the contents of the Working Directory after running.
# This saves disk space, but any additional preprocessing or analysis will have to be completely re-run.
remove_working_dir: True
log_directory:
# Whether to write log details of the pipeline run to the logging files.
run_logging: True
path: /logs
crash_log_directory:
# Directory where CPAC should write crash logs.
path: /crash
system_config:
# Select Off if you intend to run CPAC on a single machine.
# If set to On, CPAC will attempt to submit jobs through the job scheduler / resource manager selected below.
on_grid:
run: Off
# Sun Grid Engine (SGE), Portable Batch System (PBS), or Simple Linux Utility for Resource Management (SLURM).
# Only applies if you are running on a grid or compute cluster.
resource_manager: SGE
SGE:
# SGE Parallel Environment to use when running CPAC.
# Only applies when you are running on a grid or compute cluster using SGE.
parallel_environment: mpi_smp
# SGE Queue to use when running CPAC.
# Only applies when you are running on a grid or compute cluster using SGE.
queue: all.q
# The maximum amount of memory each participant's workflow can allocate.
# Use this to place an upper bound of memory usage.
# - Warning: 'Memory Per Participant' multiplied by 'Number of Participants to Run Simultaneously'
# must not be more than the total amount of RAM.
# - Conversely, using too little RAM can impede the speed of a pipeline run.
# - It is recommended that you set this to a value that when multiplied by
# 'Number of Participants to Run Simultaneously' is as much RAM you can safely allocate.
maximum_memory_per_participant: 1
# The maximum amount of cores (on a single machine) or slots on a node (on a cluster/grid)
# to allocate per participant.
# - Setting this above 1 will parallelize each participant's workflow where possible.
# If you wish to dedicate multiple cores to ANTS-based anatomical registration (below),
# this value must be equal or higher than the amount of cores provided to ANTS.
# - The maximum number of cores your run can possibly employ will be this setting multiplied
# by the number of participants set to run in parallel (the 'Number of Participants to Run
# Simultaneously' setting).
max_cores_per_participant: 1
# The number of cores to allocate to ANTS-based anatomical registration per participant.
# - Multiple cores can greatly speed up this preprocessing step.
# - This number cannot be greater than the number of cores per participant.
num_ants_threads: 1
# The number of cores to allocate to processes that use OpenMP.
num_OMP_threads: 1
# The number of participant workflows to run at the same time.
# - The maximum number of cores your run can possibly employ will be this setting
# multiplied by the number of cores dedicated to each participant (the 'Maximum Number of Cores Per Participant' setting).
num_participants_at_once: 1
# Full path to the FSL version to be used by CPAC.
# If you have specified an FSL path in your .bashrc file, this path will be set automatically.
FSLDIR: /usr/share/fsl/5.0
Amazon-AWS:
# If setting the 'Output Directory' to an S3 bucket, insert the path to your AWS credentials file here.
aws_output_bucket_credentials:
# Enable server-side 256-AES encryption on data to the S3 bucket
s3_encryption: False
Debugging:
# Verbose developer messages.
verbose: Off
# PREPROCESSING
# -------------
surface_analysis:
# Will run Freesurfer for surface-based analysis. Will output traditional Freesurfer derivatives.
# If you wish to employ Freesurfer outputs for brain masking or tissue segmentation in the voxel-based pipeline,
# select those 'Freesurfer-' labeled options further below in anatomical_preproc.
freesurfer:
run: Off
# Add extra arguments to recon-all command
reconall_args: None
# Run ABCD-HCP post FreeSurfer and fMRISurface pipeline
post_freesurfer:
run: Off
subcortical_gray_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferSubcorticalLabelTableLut.txt
freesurfer_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferAllLut.txt
surf_atlas_dir: /opt/dcan-tools/pipeline/global/templates/standard_mesh_atlases
gray_ordinates_dir: /opt/dcan-tools/pipeline/global/templates/91282_Greyordinates
gray_ordinates_res: 2
high_res_mesh: 164
low_res_mesh: 32
fmri_res: 2
smooth_fwhm: 2
longitudinal_template_generation:
# If you have multiple T1w's, you can generate your own run-specific custom
# T1w template to serve as an intermediate to the standard template for
# anatomical registration.
# This runs before the main pipeline as it requires multiple T1w sessions
# at once.
run: Off
# Freesurfer longitudinal template algorithm using FSL FLIRT
# Method to average the dataset at each iteration of the template creation
# Options: median, mean or std
average_method: median
# Degree of freedom for FLIRT in the template creation
# Options: 12 (affine), 9 (traditional), 7 (global rescale) or 6 (rigid body)
dof: 12
# Interpolation parameter for FLIRT in the template creation
# Options: trilinear, nearestneighbour, sinc or spline
interp: trilinear
# Cost function for FLIRT in the template creation
# Options: corratio, mutualinfo, normmi, normcorr, leastsq, labeldiff or bbr
cost: corratio
# Number of threads used for one run of the template generation algorithm
thread_pool: 2
# Threshold of transformation distance to consider that the loop converged
# (-1 means numpy.finfo(np.float64).eps and is the default)
convergence_threshold: -1
anatomical_preproc:
run: On
run_t2: Off
# Non-local means filtering via ANTs DenoiseImage
non_local_means_filtering:
# this is a fork option
run: [Off]
# options: 'Gaussian' or 'Rician'
noise_model: 'Gaussian'
# N4 bias field correction via ANTs
n4_bias_field_correction:
# this is a fork option
run: [Off]
# An integer to resample the input image to save computation time. Shrink factors <= 4 are commonly used.
shrink_factor: 2
# Bias field correction based on square root of T1w * T2w
t1t2_bias_field_correction:
run: Off
BiasFieldSmoothingSigma: 5
acpc_alignment:
run: Off
# Run ACPC alignment before non-local means filtering or N4 bias
# correction
run_before_preproc: True
# ACPC size of brain in z-dimension in mm.
# Default: 150mm for human data.
brain_size: 150
# Choose a tool to crop the FOV in ACPC alignment.
# Using FSL's robustfov or flirt command.
# Default: robustfov for human data, flirt for monkey data.
FOV_crop: robustfov
# ACPC Target
# options: 'brain' or 'whole-head'
# note: 'brain' requires T1w_brain_ACPC_template below to be populated
acpc_target: 'whole-head'
# ACPC aligned template
T1w_ACPC_template: /usr/share/fsl/5.0/data/standard/MNI152_T1_1mm.nii.gz
T1w_brain_ACPC_template: None
T2w_ACPC_template: None
T2w_brain_ACPC_template: None
brain_extraction:
run: On
# using: ['3dSkullStrip', 'BET', 'UNet', 'niworkflows-ants', 'FreeSurfer-ABCD', 'FreeSurfer-BET-Tight', 'FreeSurfer-BET-Loose']
# this is a fork option
using: ['3dSkullStrip']
# option parameters
AFNI-3dSkullStrip:
# Output a mask volume instead of a skull-stripped volume. The mask volume containes 0 to 6, which represents voxel's postion. If set to True, C-PAC will use this output to generate anatomical brain mask for further analysis.
mask_vol: False
# Set the threshold value controlling the brain vs non-brain voxels. Default is 0.6.
shrink_factor: 0.6
# Vary the shrink factor at every iteration of the algorithm. This prevents the likelihood of surface getting stuck in large pools of CSF before reaching the outer surface of the brain. Default is On.
var_shrink_fac: True
# The shrink factor bottom limit sets the lower threshold when varying the shrink factor. Default is 0.4, for when edge detection is used (which is On by default), otherwise the default value is 0.65.
shrink_factor_bot_lim: 0.4
# Avoids ventricles while skullstripping.
avoid_vent: True
# Set the number of iterations. Default is 250.The number of iterations should depend upon the density of your mesh.
n_iterations: 250
# While expanding, consider the voxels above and not only the voxels below
pushout: True
# Perform touchup operations at the end to include areas not covered by surface expansion.
touchup: True
# Give the maximum number of pixels on either side of the hole that can be filled. The default is 10 only if 'Touchup' is On - otherwise, the default is 0.
fill_hole: 10
# Perform nearest neighbor coordinate interpolation every few iterations. Default is 72.
NN_smooth: 72
# Perform final surface smoothing after all iterations. Default is 20.
smooth_final: 20
# Avoid eyes while skull stripping. Default is On.
avoid_eyes: True
# Use edge detection to reduce leakage into meninges and eyes. Default is On.
use_edge: True
# Speed of expansion.
exp_frac: 0.1
# Perform aggressive push to edge. This might cause leakage. Default is Off.
push_to_edge: False
# Use outer skull to limit expansion of surface into the skull in case of very strong shading artifacts. Use this only if you have leakage into the skull.
use_skull: Off
# Percentage of segments allowed to intersect surface. It is typically a number between 0 and 0.1, but can include negative values (which implies no testing for intersection).
perc_int: 0
# Number of iterations to remove intersection problems. With each iteration, the program automatically increases the amount of smoothing to get rid of intersections. Default is 4.
max_inter_iter: 4
# Multiply input dataset by FAC if range of values is too small.
fac: 1
# Blur dataset after spatial normalization. Recommended when you have lots of CSF in brain and when you have protruding gyri (finger like). If so, recommended value range is 2-4. Otherwise, leave at 0.
blur_fwhm: 0
# Set it as True if processing monkey data with AFNI
monkey: False
FSL-BET:
# Set the threshold value controling the brain vs non-brain voxels, default is 0.5
frac: 0.5
# Mask created along with skull stripping. It should be `On`, if selected functionalMasking : ['Anatomical_Refined'] and `FSL` as skull-stripping method.
mask_boolean: On
# Mesh created along with skull stripping
mesh_boolean: Off
# Create a surface outline image
outline: Off
# Add padding to the end of the image, improving BET.Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
padding: Off
# Integer value of head radius
radius: 0
# Reduce bias and cleanup neck. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
reduce_bias: Off
# Eyes and optic nerve cleanup. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
remove_eyes: Off
# Robust brain center estimation. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
robust: Off
# Create a skull image
skull: Off
# Gets additional skull and scalp surfaces by running bet2 and betsurf. This is mutually exclusive with reduce_bias, robust, padding, remove_eyes
surfaces: Off
# Apply thresholding to segmented brain image and mask
threshold: Off
# Vertical gradient in fractional intensity threshold (-1,1)
vertical_gradient : 0.0
UNet:
# UNet model
unet_model : s3://fcp-indi/resources/cpac/resources/Site-All-T-epoch_36.model
niworkflows-ants:
# Template to be used during niworkflows-ants.
# It is not necessary to change this path unless you intend to use a non-standard template.
# niworkflows-ants Brain extraction template
template_path : /ants_template/oasis/T_template0.nii.gz
# niworkflows-ants probability mask
mask_path : /ants_template/oasis/T_template0_BrainCerebellumProbabilityMask.nii.gz
# niworkflows-ants registration mask (can be optional)
regmask_path : /ants_template/oasis/T_template0_BrainCerebellumRegistrationMask.nii.gz
FreeSurfer-BET:
# Template to be used for FreeSurfer-BET brain extraction in CCS-options pipeline
T1w_brain_template_mask_ccs: /ccs_template/MNI152_T1_1mm_first_brain_mask.nii.gz
segmentation:
# Automatically segment anatomical images into white matter, gray matter,
# and CSF based on prior probability maps.
run: On
tissue_segmentation:
# using: ['FSL-FAST', 'Template_Based', 'ANTs_Prior_Based', 'FreeSurfer']
# this is a fork point
using: ['FSL-FAST']
# option parameters
FSL-FAST:
thresholding:
# thresholding of the tissue segmentation probability maps
# options: 'Auto', 'Custom'
use: 'Auto'
Custom:
# Set the threshold value for the segmentation probability masks (CSF, White Matter, and Gray Matter)
# The values remaining will become the binary tissue masks.
# A good starting point is 0.95.
# CSF (cerebrospinal fluid) threshold.
CSF_threshold_value : 0.95
# White matter threshold.
WM_threshold_value : 0.95
# Gray matter threshold.
GM_threshold_value : 0.95
use_priors:
# Use template-space tissue priors to refine the binary tissue masks generated by segmentation.
run: On
# Full path to a directory containing binarized prior probability maps.
# These maps are included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use non-standard priors.
priors_path: $FSLDIR/data/standard/tissuepriors/2mm
# Full path to a binarized White Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
WM_path: $priors_path/avg152T1_white_bin.nii.gz
# Full path to a binarized Gray Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
GM_path: $priors_path/avg152T1_gray_bin.nii.gz
# Full path to a binarized CSF prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
CSF_path: $priors_path/avg152T1_csf_bin.nii.gz
Template_Based:
# These masks should be in the same space of your registration template, e.g. if
# you choose 'EPI Template' , below tissue masks should also be EPI template tissue masks.
#
# Options: ['T1_Template', 'EPI_Template']
template_for_segmentation: ['T1_Template']
# These masks are included as part of the 'Image Resource Files' package available
# on the Install page of the User Guide.
# Full path to a binarized White Matter mask.
WHITE: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_white_bin.nii.gz
# Full path to a binarized Gray Matter mask.
GRAY: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_gray_bin.nii.gz
# Full path to a binarized CSF mask.
CSF: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_csf_bin.nii.gz
ANTs_Prior_Based:
# Generate white matter, gray matter, CSF masks based on antsJointLabelFusion
# ANTs Prior-based Segmentation workflow that has shown optimal results for non-human primate data.
# The atlas image assumed to be used in ANTs Prior-based Segmentation.
template_brain_list :
- /monkey_seg/templates/JointLabelCouncil/MacaqueYerkes19_T1w_0.5mm/T1w_brain.nii.gz
- /monkey_seg/templates/JointLabelCouncil/J_Macaque_11mo_atlas_nACQ_194x252x160space_0.5mm/T1w_brain.nii.gz
# The atlas segmentation images.
# For performing ANTs Prior-based segmentation method
# the number of specified segmentations should be identical to the number of atlas brain image sets.
# eg.
# ANTs_prior_seg_template_brain_list :
# - atlas1.nii.gz
# - atlas2.nii.gz
# ANTs_prior_seg_template_segmentation_list:
# - segmentation1.nii.gz
# - segmentation1.nii.gz
template_segmentation_list:
- /monkey_seg/templates/JointLabelCouncil/MacaqueYerkes19_T1w_0.5mm/Segmentation.nii.gz
- /monkey_seg/templates/JointLabelCouncil/J_Macaque_11mo_atlas_nACQ_194x252x160space_0.5mm/Segmentation.nii.gz
# Label values corresponding to CSF/GM/WM in atlas file
# It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
# https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/AnatomicalROI/FreeSurferColorLUT
# Label values corresponding to CSF in multiatlas file
CSF_label : [24]
# Label values corresponding to Gray Matter in multiatlas file
GM_label : [3, 42]
# Label values corresponding to White Matter in multiatlas file
WM_label : [2, 41]
FreeSurfer:
# Use mri_binarize --erode option to erode segmentation masks
erode: 0
# Label values corresponding to CSF in FreeSurfer aseg segmentation file
CSF_label : [24]
# Label values corresponding to Gray Matter in FreeSurfer aseg segmentation file
GM_label : [3, 42]
# Label values corresponding to White Matter in FreeSurfer aseg segmentation file
WM_label : [2, 41]