I reproduced the error on the same dataset, and it looks like the priors are causing this problem. Did you specifically want to use priors?
It would be better to apply the same preprocessing configurations to all the subjects, but if not using priors can lead to a reasonable output, I think not using priors is cool, I will try it on the data to see if there are any further questions, thanks for your help!
by the way, do I just need to change use_priors run: On to use_priors run: Off or I need to make more changes?
Yes, that’s all you have to change!
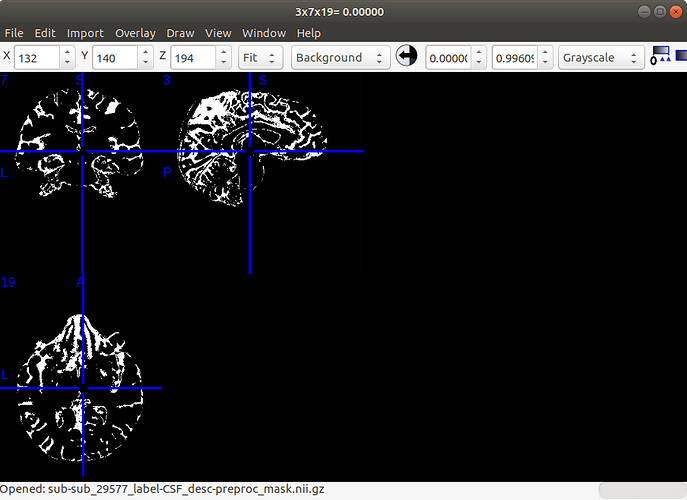
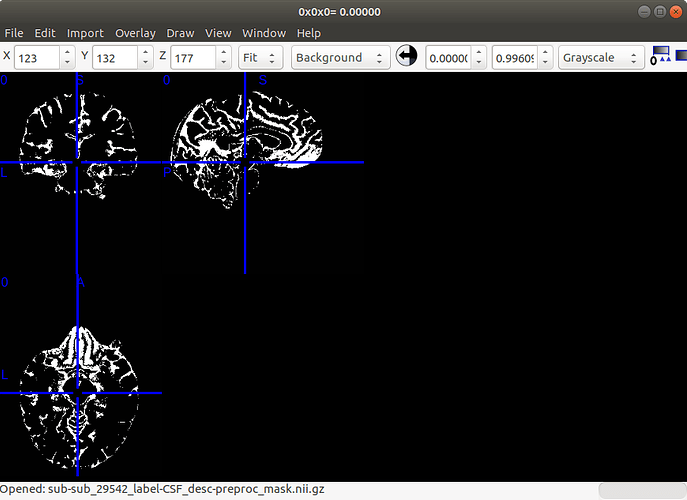
Hi, sorry for the late reply, I followed your advice and subject-29576 can be preprocessed perfectly, however, when I apply the same pipeline to other subjects, although the CSF file is alright the ‘ValueError: Number of time points’ still remains.
Below is the CSF mask result:
And here is the log files:
pypeline.txt (361.7 KB)
29542_pypeline.txt (362.5 KB)
29576_pypeline.txt (809.7 KB)
29577_pypeline.txt (362.3 KB)
Could you please help me to find out which part goes wrong?
Thank you for your time!
After running some tests, it looks like the lateral ventricle mask may be too stringent for some of the subjects, which is why this error appears to be data dependent. The error disappeared when I ran this pipeline without the lateral mask. Omitting the mask would be a less conservative approach to extracting the CSF mask, but it seems like it isn’t compatible with the dataset. You can just replace the path to the mask with None. Let me know if you have any questions!
Hi @ekenneally,
I’m trying to do the same thing: extracting time series using CPAC as the processing pipeline for ABIDE 2. I used the default pipeline and configured it to produce the same time series for the atlases I have in ABIDE 1. Previously, to ensure my configuration file was correct, I tested it on ABIDE 1. However, when I compared my time series with those from ABIDE 1, I found that they were not the same—not even approximately.
What can I do to generate the same results?
Hi @ichkifa, thanks for reaching out! Are you able to share your configuration file so I can look into this further?
%YAML 1.1
---
pipeline_setup:
pipeline_name: cpacImanitaFinal1
output_directory:
path: /outputs/output
source_outputs_dir: None
pull_source_once: True
write_func_outputs: True
write_debugging_outputs: True
output_tree: "default"
# Quality control outputs
quality_control:
# Generate quality control pages containing preprocessing and derivative outputs.
generate_quality_control_images: False
generate_xcpqc_files: False
working_directory:
path: /work
remove_working_dir: True
log_directory:
run_logging: True
path: /outputs/log
graphviz:
# Configuration for a graphviz visualization of the entire workflow. See https://fcp-indi.github.io/docs/developer/nodes#CPAC.pipeline.nipype_pipeline_engine.Workflow.write_graph for details about the various options
entire_workflow:
# Whether to generate the graph visualization
generate: Off
# Options: [orig, hierarchical, flat, exec, colored]
graph2use: []
# Options: [svg, png]
format: []
# The node name will be displayed in the form `nodename (package)` when On or `nodename.Class.package` when Off
simple_form: On
crash_log_directory:
path: /outputs/crash
system_config:
fail_fast: Off
random_seed:
# The maximum amount of memory each participant's workflow can allocate.
# Use this to place an upper bound of memory usage.
# - Warning: 'Memory Per Participant' multiplied by 'Number of Participants to Run Simultaneously'
# must not be more than the total amount of RAM.
# - Conversely, using too little RAM can impede the speed of a pipeline run.
# - It is recommended that you set this to a value that when multiplied by
# 'Number of Participants to Run Simultaneously' is as much RAM you can safely allocate.
maximum_memory_per_participant: 1
# Prior to running a pipeline C-PAC makes a rough estimate of a worst-case-scenario maximum concurrent memory usage with high-resoltion data, raising an exception describing the recommended minimum memory allocation for the given configuration.
# Turning this option off will allow pipelines to run without allocating the recommended minimum, allowing for more efficient runs at the risk of out-of-memory crashes (use at your own risk)
raise_insufficient: On
# A callback.log file from a previous run can be provided to estimate memory usage based on that run.
observed_usage:
# Path to callback log file with previously observed usage.
# Can be overridden with the commandline flag `--runtime_usage`.
callback_log:
# Percent. E.g., `buffer: 10` would estimate 1.1 * the observed memory usage from the callback log provided in "usage".
# Can be overridden with the commandline flag `--runtime_buffer`.
buffer: 10
max_cores_per_participant: 1
num_ants_threads: 1
num_OMP_threads: 1
num_participants_at_once: 1
FSLDIR: FSLDIR
Amazon-AWS:
# If setting the 'Output Directory' to an S3 bucket, insert the path to your AWS credentials file here.
aws_output_bucket_credentials:
# Enable server-side 256-AES encryption on data to the S3 bucket
s3_encryption: False
Debugging:
# Verbose developer messages.
verbose: On
##################################################
anatomical_preproc:
run: On
brain_extraction:
run: On
using: ['3dSkullStrip']
functional_preproc:
run: On
truncation:
start_tr: 0
stop_tr: None
func_masking:
run: On
using: ['AFNI']
slice_timing_correction:
# Interpolate voxel time courses so they are sampled at the same time points.
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [On]
# use specified slice time pattern rather than one in header
tpattern: None
motion_estimates_and_correction:
run: On
motion_estimates:
# calculate motion statistics BEFORE slice-timing correction
calculate_motion_first: Off
# calculate motion statistics AFTER motion correction
calculate_motion_after: On
motion_correction:
# using: ['3dvolreg', 'mcflirt']
# Forking is currently broken for this option.
# Please use separate configs if you want to use each of 3dvolreg and mcflirt.
# Follow https://github.com/FCP-INDI/C-PAC/issues/1935 to see when this issue is resolved.
using: ['3dvolreg']
# option parameters
AFNI-3dvolreg:
# This option is useful when aligning high-resolution datasets that may need more alignment than a few voxels.
functional_volreg_twopass: On
# Choose motion correction reference. Options: mean, median, selected_volume, fmriprep_reference
motion_correction_reference: ['mean']
# Choose motion correction reference volume
motion_correction_reference_volume: 0
generate_func_mean:
# Generate mean functional image
run: On
normalize_func:
# Normalize functional image
run: On
coreg_prep:
# Generate sbref
run: On
segmentation:
# Automatically segment anatomical images into white matter, gray matter,
# and CSF based on prior probability maps.
run: On
tissue_segmentation:
# using: ['FSL-FAST', 'Template_Based', 'ANTs_Prior_Based', 'FreeSurfer']
# this is a fork point
using: ['FSL-FAST']
# option parameters
FSL-FAST:
thresholding:
# thresholding of the tissue segmentation probability maps
# options: 'Auto', 'Custom'
use: 'Auto'
Custom:
# Set the threshold value for the segmentation probability masks (CSF, White Matter, and Gray Matter)
# The values remaining will become the binary tissue masks.
# A good starting point is 0.95.
# CSF (cerebrospinal fluid) threshold.
CSF_threshold_value : 0.96
# White matter threshold.
WM_threshold_value : 0.96
# Gray matter threshold.
GM_threshold_value : 0.7
use_priors:
# Use template-space tissue priors to refine the binary tissue masks generated by segmentation.
run: On
# Full path to a directory containing binarized prior probability maps.
# These maps are included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use non-standard priors.
priors_path: $FSLDIR/data/standard/tissuepriors/2mm
# Full path to a binarized White Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
WM_path: $priors_path/avg152T1_white_bin.nii.gz
# Full path to a binarized Gray Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
GM_path: $priors_path/avg152T1_gray_bin.nii.gz
# Full path to a binarized CSF prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
CSF_path: $priors_path/avg152T1_csf_bin.nii.gz
nuisance_corrections:
1-ICA-AROMA:
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [Off]
# Types of denoising strategy:
# nonaggr: nonaggressive-partial component regression
# aggr: aggressive denoising
denoising_type: nonaggr
2-nuisance_regression:
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [On]
# this is not a fork point
# Run nuisance regression in native or template space
# - If set to template, will use the brain mask configured in
# ``functional_preproc: func_masking: FSL_AFNI: brain_mask``
# - If ``registration_workflows: functional_registration: func_registration_to_template: apply_trasnform: using: single_step_resampling_from_stc``, this must be set to template
space: native
# switch to Off if nuisance regression is off and you don't want to write out the regressors
create_regressors: On
# Select which nuisance signal corrections to apply
Regressors:
- Name: 'filt_global'
Motion:
include_delayed: true
include_squared: true
include_delayed_squared: true
aCompCor:
summary:
method: DetrendPC
components: 5
tissues:
- WhiteMatter
- CerebrospinalFluid
extraction_resolution: 2
CerebrospinalFluid:
summary: Mean
extraction_resolution: 2
erode_mask: true
GlobalSignal:
summary: Mean
PolyOrt:
degree: 2
Bandpass:
bottom_frequency: 0.01
top_frequency: 0.1
method: default
# Standard Lateral Ventricles Binary Mask
# used in CSF mask refinement for CSF signal-related regressions
lateral_ventricles_mask: $FSLDIR/data/atlases/HarvardOxford/HarvardOxford-lateral-ventricles-thr25-2mm.nii.gz
# Whether to run frequency filtering before or after nuisance regression.
# Options: 'After' or 'Before'
bandpass_filtering_order: 'After'
# Process and refine masks used to produce regressors and time series for
# regression.
regressor_masks:
erode_anatomical_brain_mask:
# Erode binarized anatomical brain mask. If choosing True, please also set regressor_masks['erode_csf']['run']: True; anatomical_preproc['brain_extraction']['using']: niworkflows-ants.
run: Off
# Target volume ratio, if using erosion.
# Default proportion is None for anatomical brain mask.
# If using erosion, using both proportion and millimeters is not recommended.
brain_mask_erosion_prop:
# Erode brain mask in millimeters, default for brain mask is 30 mm
# Brain erosion default is using millimeters.
brain_mask_erosion_mm: 30
# Erode binarized brain mask in millimeter
brain_erosion_mm:
erode_csf:
# Erode binarized csf tissue mask.
run: Off
# Target volume ratio, if using erosion.
# Default proportion is None for cerebrospinal fluid mask.
# If using erosion, using both proportion and millimeters is not recommended.
csf_erosion_prop:
# Erode cerebrospinal fluid mask in millimeters, default for cerebrospinal fluid is 30mm
# Cerebrospinal fluid erosion default is using millimeters.
csf_mask_erosion_mm: 30
# Erode binarized cerebrospinal fluid mask in millimeter
csf_erosion_mm:
erode_wm:
# Erode WM binarized tissue mask.
run: Off
# Target volume ratio, if using erosion.
# Default proportion is 0.6 for white matter mask.
# If using erosion, using both proportion and millimeters is not recommended.
# White matter erosion default is using proportion erosion method when use erosion for white matter.
wm_erosion_prop: 0.6
# Erode white matter mask in millimeters, default for white matter is None
wm_mask_erosion_mm:
# Erode binarized white matter mask in millimeters
wm_erosion_mm:
erode_gm:
# Erode gray matter binarized tissue mask.
run: Off
# Target volume ratio, if using erosion.
# If using erosion, using both proportion and millimeters is not recommended.
gm_erosion_prop: 0.6
# Erode gray matter mask in millimeters
gm_mask_erosion_mm:
# Erode binarized gray matter mask in millimeters
gm_erosion_mm:
post_processing:
z-scoring:
run: Off
registration_workflows:
anatomical_registration:
run: On
resolution_for_anat: 2mm
T1w_brain_template: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain.nii.gz
T1w_template: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}.nii.gz
T1w_brain_template_mask: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask.nii.gz
reg_with_skull: True
registration:
using: ['ANTS']
FSL-FNIRT:
fnirt_config: T1_2_MNI152_2mm
ref_mask: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask_symmetric_dil.nii.gz
functional_registration:
coregistration:
# functional (BOLD/EPI) registration to anatomical (structural/T1)
run: On
func_input_prep:
# Choose whether to use functional brain or skull as the input to functional-to-anatomical registration
reg_with_skull: Off
# Choose whether to use the mean of the functional/EPI as the input to functional-to-anatomical registration or one of the volumes from the functional 4D timeseries that you choose.
# input: ['Mean_Functional', 'Selected_Functional_Volume', 'fmriprep_reference']
input: ['Mean_Functional']
Selected Functional Volume:
# Only for when 'Use as Functional-to-Anatomical Registration Input' is set to 'Selected Functional Volume'.
#Input the index of which volume from the functional 4D timeseries input file you wish to use as the input for functional-to-anatomical registration.
func_reg_input_volume: 0
boundary_based_registration:
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [On]
# Standard FSL 5.0 Scheduler used for Boundary Based Registration.
# It is not necessary to change this path unless you intend to use non-standard MNI registration.
bbr_schedule: $FSLDIR/etc/flirtsch/bbr.sch
func_registration_to_template:
# these options modify the application (to the functional data), not the calculation, of the
# T1-to-template and EPI-to-template transforms calculated earlier during registration
# apply the functional-to-template (T1 template) registration transform to the functional data
run: On
output_resolution:
# The resolution (in mm) to which the preprocessed, registered functional timeseries outputs are written into.
# NOTE:
# selecting a 1 mm or 2 mm resolution might substantially increase your RAM needs- these resolutions should be selected with caution.
# for most cases, 3 mm or 4 mm resolutions are suggested.
# NOTE:
# this also includes the single-volume 3D preprocessed functional data,
# such as the mean functional (mean EPI) in template space
func_preproc_outputs: 3mm
# The resolution (in mm) to which the registered derivative outputs are written into.
# NOTE:
# this is for the single-volume functional-space outputs (i.e. derivatives)
# thus, a higher resolution may not result in a large increase in RAM needs as above
func_derivative_outputs: 3mm
target_template:
# choose which template space to transform derivatives towards
# using: ['T1_template', 'EPI_template']
# this is a fork point
# NOTE:
# this will determine which registration transform to use to warp the functional
# outputs and derivatives to template space
using: ['T1_template']
T1_template:
# Standard Skull Stripped Template. Used as a reference image for functional registration.
# This can be different than the template used as the reference/fixed for T1-to-template registration.
T1w_brain_template_funcreg: $FSLDIR/data/standard/MNI152_T1_3mm_brain.nii.gz
# Standard Anatomical Brain Image with Skull.
# This can be different than the template used as the reference/fixed for T1-to-template registration.
T1w_template_funcreg: $FSLDIR/data/standard/MNI152_T1_3mm.nii.gz
# Template to be used during registration.
# It is not necessary to change this path unless you intend to use a non-standard template.
T1w_brain_template_mask_funcreg: $FSLDIR/data/standard/MNI152_T1_3mm_brain_mask.nii.gz
# a standard template for resampling if using float resolution
T1w_template_for_resample: $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz
FNIRT_pipelines:
# Interpolation method for writing out transformed functional images.
# Possible values: trilinear, sinc, spline
interpolation: sinc
# Identity matrix used during FSL-based resampling of functional-space data throughout the pipeline.
# It is not necessary to change this path unless you intend to use a different template.
identity_matrix: $FSLDIR/etc/flirtsch/ident.mat
timeseries_extraction:
run: On
tse_roi_paths:
/cpac_templates/CC400.nii.gz: Avg
/cpac_templates/CC200.nii.gz: Avg
/cpac_templates/aal_mask_pad.nii.gz: Avg
realignment: 'ROI_to_func'
connectivity_matrix:
# Create a connectivity matrix from timeseries data
# Options:
# ['AFNI', 'Nilearn', 'ndmg']
using:
- Nilearn
measure:
- Pearson
# PACKAGE INTEGRATIONS
# --------------------
PyPEER:
# Training of eye-estimation models. Commonly used for movies data/naturalistic viewing.
run: Off
# PEER scan names to use for training
# Example: ['peer_run-1', 'peer_run-2']
eye_scan_names: []
# Naturalistic viewing data scan names to use for eye estimation
# Example: ['movieDM']
data_scan_names: []
# Template-space eye mask
eye_mask_path: $FSLDIR/data/standard/MNI152_T1_${func_resolution}_eye_mask.nii.gz
# PyPEER Stimulus File Path
# This is a file describing the stimulus locations from the calibration sequence.
stimulus_path: None
minimal_nuisance_correction:
# PyPEER Minimal nuisance regression
# Note: PyPEER employs minimal preprocessing - these choices do not reflect what runs in the main pipeline.
# PyPEER uses non-nuisance-regressed data from the main pipeline.
# Global signal regression (PyPEER only)
peer_gsr: True
# Motion scrubbing (PyPEER only)
peer_scrub: False
# Motion scrubbing threshold (PyPEER only)
scrub_thresh: 0.2
Hi @ichkifa,
Thank you! Can I ask you to clarify whether you were able to replicate the preprocessing from ABIDE I, but haven’t been able replicate it for ABIDE II, or if you’re comparing ABIDE II outputs to ABIDE I outputs?
Looking at the ABIDE II documentation, it appears that there are a handful of subjects that have data in common with ABIDE I, but not all of them. Since the timeseries are therefore coming from different participants, and may also have been produced using a range of C-PAC versions and pipeline configs, they won’t be the same when comparing ABIDE I/II.
Hello @tamsinrogers,
Thank you for your message!
I am currently working on reproducing the time series of ABIDE I from the raw fMRI images, using C-PAC with the same configuration that was used for ABIDE preprocessed.
The goal is to validate my configuration by comparing the time series I obtain with those already published for ABIDE I. This will ensure that my configuration is correct before using it to process the ABIDE II data.
Best regards,
Hi @ichkifa,
Thanks for the clarification! Our team is reproducing on our end and will get back to you shortly.
Hi @ichkifa,
Our team is still working to replicate this issue, which will hopefully help us move forward with answering your question. I will get back to you as soon as I have some more information on how to move forward with this!
Thank you for the update. I look forward to your feedback. Let me know when you have more information!
Best regards,
Hello @ichkifa,
I work with Tamsin on the C-PAC team and have been following this thread. We worked on replicating your run for the purpose of scoping just how different the outputs are. As expected, many of the intermediate outputs (T1w in template, masks, etc.) are not too different, but the final denoised timeseries in template space sees a more dramatic change across the versions.
One of our design principles is for C-PAC to remain consistent over time, with regression testing ensuring outputs remain stable across versions (typically Pearson r ≥ 0.98). However, we are looking at versions that are roughly 10 years apart, so it’s not surprising that larger differences emerged. These are most likely due to:
- A few intentional but impactful pipeline updates over the years (documented in the release notes along the way).
- Small drifts from software dependency changes, accumulating over time. However, over the past 6 or 7 years, the software dependency versions have been locked and documented (with those versions being established in each release’s container), reducing this effect. However, an important and nearly impossible example of this drift to track is the core numerical processing libraries, like
glibc, that is used by many of the processing tools and does influence the reproducibility of floating point operations. - Intermediate outputs having high but not perfect correlations (e.g., ~0.95), which compound as they propagate through multiple processing steps. If you examine the earlier outputs of the pipeline (for example, T1w brain masking and registration to template), you will find they remain largely stable (r ≥ 0.92-0.95). However, with the non-deterministic nature of tissue segmentation and nonlinear registration, the slight differences in these early outputs then go on to steps like coregistration, nuisance denoising (particularly things like CSF signal regression and aCompCor) and this results in an outsized difference in the final template-space denoised timeseries.
If you are still concerned about benchmarking against the original ABIDE Preprocessed results, let us know and we can help you determine what sort of criteria or goals/aims you are looking to achieve. If you can provide some more information as to why you originally wanted to benchmark the outputs, we can have a discussion as to what data are appropriate for your experiment.
Hi @sgiavasis
Thank you very much for your response and follow-up.
As I am not a specialist in neuroimaging preprocessing but a researcher working on autism, I would like to clarify whether, based on what you explained, it is indeed impossible to obtain exactly the same results as those published due to updates in the pipeline.
Additionally, I have noticed differences in the number of time points in the published time series compared to the raw data. For example, for the CMU_a site, the JSON file and the 4th dimension of the raw fMRI data indicate 240 observations, but the published time series contain only 236. Similarly, for Caltech, the published series have 152 observations, whereas the JSON file and fMRI images indicate 156. The same discrepancy appears for other sites as well.
Could you explain why these differences exist? Which specific time points are removed or added, and based on what criteria?
Best regards,
Hi @ichkifa,
The discrepancy between the number of time points in the published time series compared to the raw data (-4 TRs) is due to the the pipeline configuration for the published outputs being set to drop the first 4 TRs.
startIdx: 4
This is a common practice in neuroimaging, it helps control for the warmup drift that some MR scanners have, and explains why you’re seeing raw: 240 / published: 236, raw: 156 / published: 152, etc. Essentially, cutting out the first few seconds of the scan where there might be any subject movement or machine initialization happening helps ensure a more stable overall scan.
Please let me know if I can clarify further!