Dear all,
We are trying to preprocess our BOLD data with --use-syn-sdc with the update version. The data were collected using GE 3T scanner. Here is the error message:
ValueError: Fieldmap-less (SyN) estimation was requested, but PhaseEncodingDirection information appears to be absent.
Here is our script:
bids_root_dir=bids_test5_fieldmaplessonly
subj=3172101920
nthreads=4
mem=40
container=singularity
fmriprep $bids_root_dir $bids_root_dir/derivatives
participant
–participant-label $subj
–skip-bids-validation
–md-only-boilerplate
–fs-license-file /scratch/g/jgoveas/R01_nii/baseline_mid/bids/license.txt
–fs-no-reconall
–output-spaces MNI152NLin2009cAsym:res-2
–nthreads $nthreads
–mem_mb $mem_mb
–ignore slicetiming
–ignore fieldmaps
–use-syn-sdc
-w fmriprep_work_MID
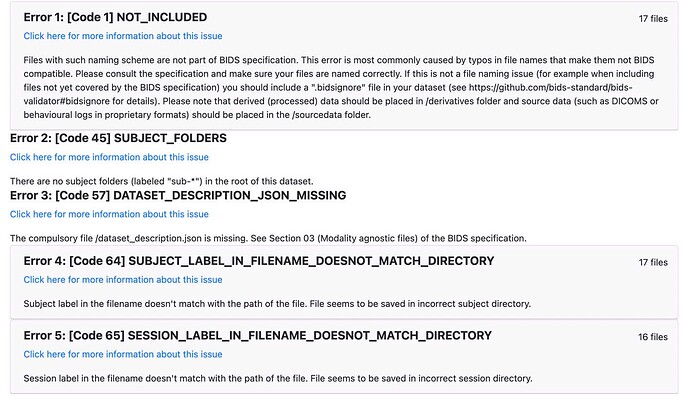
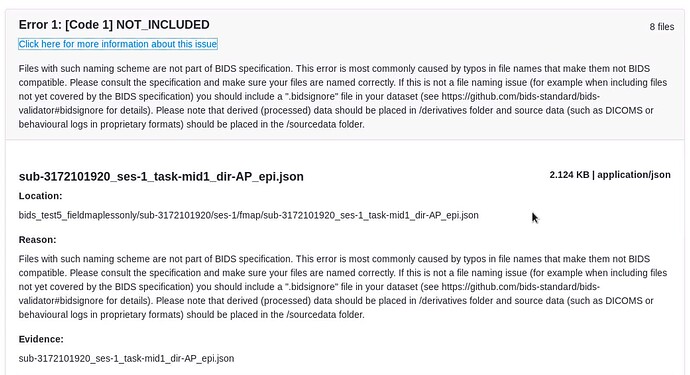
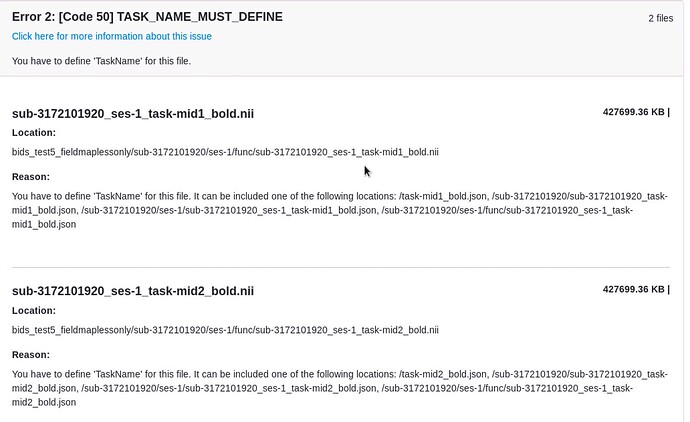
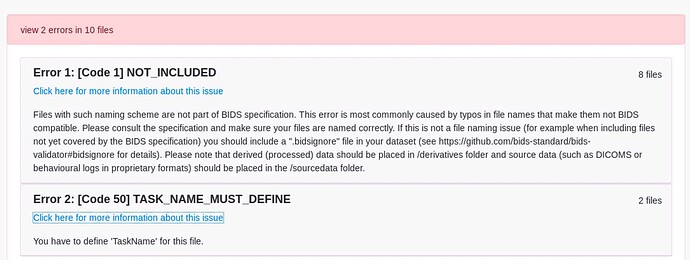
Here are our data:
anat/
-sub-3172101920_ses-1_T1w.json
-sub-3172101920_ses-1_T1w.nii
func/
-sub-3172101920_ses-1_task-mid1_bold.json
-sub-3172101920_ses-1_task-mid1_bold.nii
-sub-3172101920_ses-1_task-mid2_bold.json
-sub-3172101920_ses-1_task-mid2_bold.nii
We were able to run this successfully using v. 20.2.3. The only difference was that we added --ignore fieldmaps.
Thank you for your help!