Hi,
I added the -B arguments. The script ran successfully but it still showed “susceptibility distortion correction: none” in the report.
Here is the script
#User inputs:
bids_root_dir=/scratch/g/jgoveas/R01_nii/baseline_mid/PROJECT/bidstest5/
subj=3172101920
nthreads=4
mem=40 #gb
container=singularity #docker or singularity
#Begin:
#Convert virtual memory from gb to mb
mem=echo "${mem//[!0-9]/}"
mem_mb=echo $(((mem*1000)-5000))
export FS_LICENSE=/scratch/g/jgoveas/R01_nii/baseline_mid/bids/license.txt \
singularity run
-B /scratch/g/jgoveas/R01_nii/baseline_mid
-B /scratch/g/jgoveas/R01_nii/baseline_mid/PROJECT
-B /scratch/g/jgoveas/R01_nii/baseline_mid/PROJECT/bidstest5
–cleanenv /scratch/g/jgoveas/R01_nii/baseline_mid/fmriprep-22.0.2.simg $bids_root_dir $bids_root_dir/derivatives
participant
–participant-label $subj
–skip-bids-validation
–md-only-boilerplate
–fs-license-file /scratch/g/jgoveas/R01_nii/baseline_mid/bids/license.txt
–fs-no-reconall
–output-spaces MNI152NLin2009cAsym:res-2
–output-spaces func:res-native
–output-spaces MNI152NLin2009cAsym:res-native
–nthreads $nthreads
–mem_mb mem_mb \
--ignore slicetiming \
-w /scratch/g/jgoveas/R01_nii/baseline_mid/TOPUP_{subj}
Here is the tree hierarchy:
sub-3172101920
└── ses-1
├── anat
│ ├── sub-3172101920_ses-1_FLAIR.json
│ ├── sub-3172101920_ses-1_FLAIR.nii
│ ├── sub-3172101920_ses-1_T1w.json
│ └── sub-3172101920_ses-1_T1w.nii
├── fmap
│ ├── sub-3172101920_ses-1_acq-mid1_dir-AP_epi.json
│ ├── sub-3172101920_ses-1_acq-mid1_dir-AP_epi.nii
│ ├── sub-3172101920_ses-1_acq-mid1_dir-PA_epi.json
│ ├── sub-3172101920_ses-1_acq-mid1_dir-PA_epi.nii
│ ├── sub-3172101920_ses-1_acq-mid2_dir-AP_epi.json
│ ├── sub-3172101920_ses-1_acq-mid2_dir-AP_epi.nii
│ ├── sub-3172101920_ses-1_acq-mid2_dir-PA_epi.json
│ └── sub-3172101920_ses-1_acq-mid2_dir-PA_epi.nii
└── func
├── sub-3172101920_ses-1_task-mid1_bold.json
├── sub-3172101920_ses-1_task-mid1_bold.nii
├── sub-3172101920_ses-1_task-mid2_bold.json
└── sub-3172101920_ses-1_task-mid2_bold.nii
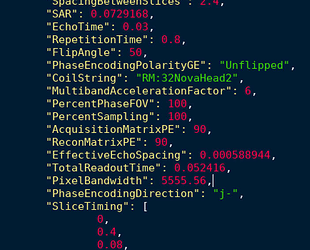
Here is one of the fmap.json
{
“Modality”: “MR”,
“MagneticFieldStrength”: 3,
“ImagingFrequency”: 127.772,
“Manufacturer”: “GE”,
“PulseSequenceName”: “epi”,
“InternalPulseSequenceName”: “EPI”,
“ManufacturersModelName”: “SIGNA Premier”,
“InstitutionName”: “MCW Premier”,
“DeviceSerialNumber”: “00000000FLORIO1X”,
“StationName”: “MCWPREM”,
“BodyPartExamined”: “BRAIN”,
“PatientPosition”: “HFS”,
“ProcedureStepDescription”: “GovGriefO1”,
“SoftwareVersions”: “27\LX\MR Software release:RX27.0_R03_1850.a”,
“MRAcquisitionType”: “2D”,
“SeriesDescription”: “SE Map PE2”,
“ProtocolName”: “Goveas_GovGriefR01_10052”,
“ScanningSequence”: “EP\RM”,
“SequenceVariant”: “NONE”,
“ScanOptions”: “SAT_GEMS\MP_GEMS\EPI_GEMS\FS”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “OTHER”],
“SeriesNumber”: 26,
“AcquisitionTime”: “10:20:1.000000”,
“AcquisitionNumber”: 1,
“TriggerDelayTime”: 14677,
“SliceThickness”: 2.4,
“SpacingBetweenSlices”: 2.4,
“SAR”: 0.20231,
“EchoTime”: 0.08,
“RepetitionTime”: 7.4,
“FlipAngle”: 90,
“PhaseEncodingPolarityGE”: “Unflipped”,
“CoilString”: “RM:32NovaHead2”,
“PercentPhaseFOV”: 100,
“PercentSampling”: 100,
“AcquisitionMatrixPE”: 90,
“ReconMatrixPE”: 128,
“EffectiveEchoSpacing”: 0.000403654,
“TotalReadoutTime”: 0.051264,
“IntendedFor”: “ses-1/func/sub-3172101920_ses-1_task-mid1_bold.nii”,
“PixelBandwidth”: 3906.25,
“PhaseEncodingDirection”: “j-”,
“SliceTiming”: [
0,
3.7,
0.123333,
3.82333,
0.246667,
3.94667,
0.37,
4.07,
0.493333,
4.19333,
0.616667,
4.31667,
0.74,
4.44,
0.863333,
4.56333,
0.986667,
4.68667,
1.11,
4.81,
1.23333,
4.93333,
1.35667,
5.05667,
1.48,
5.18,
1.60333,
5.30333,
1.72667,
5.42667,
1.85,
5.55,
1.97333,
5.67333,
2.09667,
5.79667,
2.22,
5.92,
2.34333,
6.04333,
2.46667,
6.16667,
2.59,
6.29,
2.71333,
6.41333,
2.83667,
6.53667,
2.96,
6.66,
3.08333,
6.78333,
3.20667,
6.90667,
3.33,
7.03,
3.45333,
7.15333,
3.57667,
7.27667 ],
“ImageOrientationPatientDICOM”: [
1,
-0,
0,
-0,
1,
0 ],
“InPlanePhaseEncodingDirectionDICOM”: “COL”,
“ConversionSoftware”: “dcm2niix”,
“ConversionSoftwareVersion”: “v1.0.20210317”
}
Please let me know what is missing.
Thank you for your help.