Hi all, Is it possible with nilearn to obtain volume values by ROIs from a T1 that has been preprocessed on fmriprep?
Best regard,
Raul
Hi,
Yes, it is quite possible, using a NiftiMasker, providing as mask the binary image the defines the ROI. Please take time to read
HTH,
Bertrand
Ahoi hoi @rglezgz,
just to follow up on @bthirion’s comment, depending on your data and the planned analyses, different options of nilearn.maskers could be feasible/interesting. For example, if you want to extract volume values from multiple ROIs and participants, the following example could be helpful:
# import modules
from nilearn import datasets
from nilearn.input_data import NiftiLabelsMasker
import nibabel as nb
# download VBM data from 10 example participants
vbm_exp = datasets.fetch_oasis_vbm(n_subjects=10)
# download an example atlas, here AAL
aal_atlas = datasets.fetch_atlas_aal()
# set up NiftiLabelsMasker to extract volume values from multiple ROIs (the AAL atlas)
aal_masker = NiftiLabelsMasker(labels_img=aal_atlas.maps)
# define an empty list to store volume values for each participant
vbm_exp_data = []
# loop over participants, respectively extract and storing volume values
for i,vbm_img in enumerate(vbm_exp['gray_matter_maps']):
# load the current image using nibabel
vbm_exp_img = nb.load(vbm_img)
# extract volume value per ROI from current image and append it to the list
vbm_exp_data.append(aal_masker.fit_transform(vbm_exp_img).squeeze())
# convert the list into a pandas DataFrame, assigning atlas labels as columns
df_vbm_data = pd.DataFrame(vbm_exp_data, columns=aal_atlas.labels)
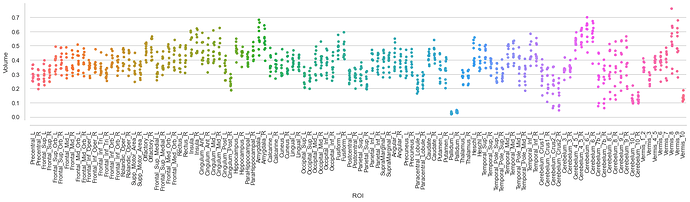
With this relatively straightforward approach, you get a DataFrame that can be utilized for statistical analyses and/or visualization. For example, showing volume values for all participants per ROI:
# import modules
import seaborn as sns
# set figure parameters
sns.set(rc={"figure.figsize":(22, 4)})
sns.set_style('whitegrid')
# create plot
g = sns.stripplot(df_vbm_data)
# set some aesthetics
sns.despine(offset=10)
# rotate labels for readability
g.set_xticklabels(labels=df_vbm_data.columns, rotation=90);
# assign axis labels
g.set_ylabel('Volume');
g.set_xlabel('ROI');
HTH, cheers, Peer
Excellent, thanks to both. @PeerHerholz that script is very helpful, but I still need to obtain the TIV values to adjust the ROI volumes.
TIV can be found in the FreeSurfer outputs /path/to/Freesurfer/sub-XX/stats/brainvol.stats or aseg.stats.
Hi @Steven, I don’t understand how to find the TIV value. I just ran the script of @PeerHerholz and I can’t find these outputs.
I want to use this script on a dataset preprocessed by fmriprep, but for T1 there is no TIV as an output.
thanks for you time
Did you run FreeSurfer as part of fmriprep? If so, you can find the TIV in the statistics text file output by FreeSurfer in the path I described in my previous post. If not, you can run recon-all from FreeSurfer on your T1, which will output the TIV (and other stats) in the text files I mentioned in my previous answer.
Thanks, @Steven. I just used --fs-no-reconall to expend less time in the fmriprep analysis. I’m running now.