Hello everyone,
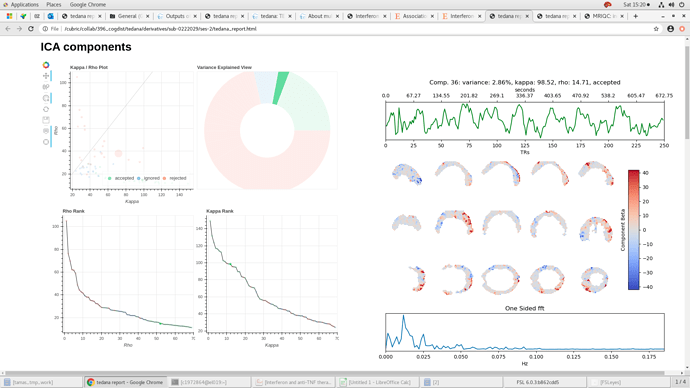
I wonder if someone has come across incomplete beta maps when using tedana. The times series data and individual bold reports from fmriprep look ok . We have a few tedana outputs like this one and, for some of those participants we got more noisy data with FD mean values above threshold, so just wondering if that could be a potential explanation.
Any suggestions would be very much appreciated
Many thanks in advance for your help.
One thing that comes to mind for me is that the masking from tedana was way too aggressive. You are saying this effect is only for some participants? Excessive motion in the timeseries shouldn’t produce this effect, I don’t think, unless it was so extreme that the data was compromised and was more artifact than signal.
Can you provide all of the sequence parameters, including field strength and echo times? If possible, screenshots of the EPI data at each echo time may also be useful - a 3slice multiplanar view near the middle of the brain perhaps?
Hello,
Thanks for your message.
So, the sequence parameters are 3T , TR 26914 ms and TEs 12.60, 30.25 and 47.9 ms.
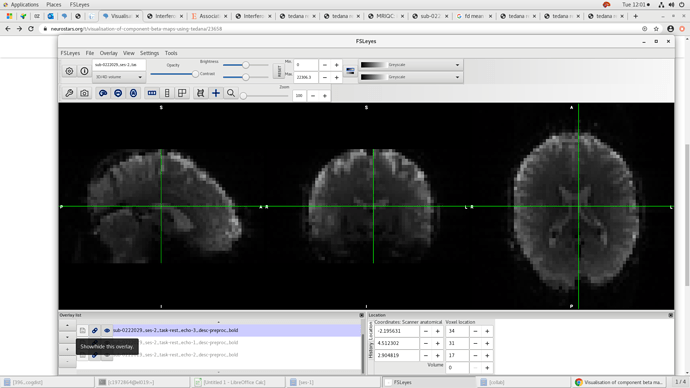
Below you can see the multiplanar view of one echo. Sorry i’m only allowed to upload one image but they all look similar.
Many thanks again for you help
Best wishes
Eva
Hmm. That is the 3rd echo, correct? It looks fine to me, but it really seems like tedana masked out those low intensity values towards the center…even though that don’t look excessively low.
tedana has a mask option (Using tedana from the command line — tedana 0.0.12+0.g863304b.dirty documentation) --mask somemaskfile.nii.gz that you may want to try. I’m not sure if fmriprep automatically gives you some mask choices, but that would be my next step. I tend to use an automask (in the AFNI 3dAutomask sense) from the first echo - includes brain, perhaps a little bit of skull, but excludes areas outside head and with extremely low signal.
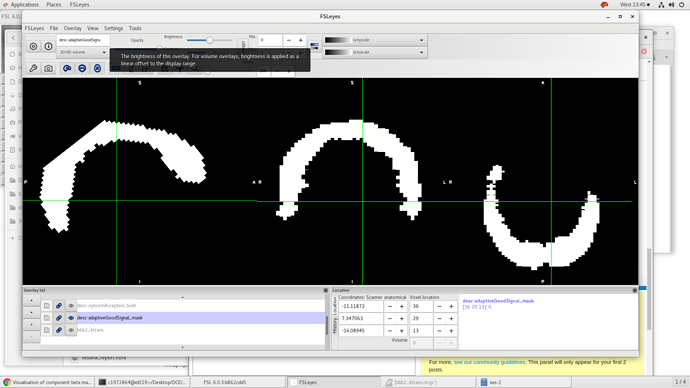
In fact, tedana should have produced some mask files - I assume those look like the beta maps there - missing a lot of brain?
Hi,
yest that’s the 3rd echo and yes the mask files also have big parts of the brain missing, please see below.
Many thanks for the suggestions, we’ll give it a try, hopefully that’ll sort out the issue
Ok, then that at least makes sense and hopefully using an explicit mask resolves this problem, though it is strange that this mask is so restrictive despite the 3rd echo looking ok. It may just be unlucky, or could be related to some of the specific cut offs used in the adaptive masking step. Good luck, and looking forward to an update. Fingers crossed.
Edit: I do wonder if the motion is extreme enough that it is dragging the mean down - but it would have to be really excessive to have that effect with that many timepoints. But who knows. Its possible that someone may reach out to ask about this data if we begin to examine the adaptive masking code, because this seems like fairly typical 3T data and it is surprising that things went wrong.
The average FD across the 3 echos is below threshold, so not sure this issue’s got anything to do with extreme motion artifacts. My colleague @foldes.andrei (Tamas) is now running the automask with AFNI, so fingers croseed. Many thanks!!
As @dowdlelt mentioned, we used to have more aggressive masking within tedana so that, if any of the echoes had too little signal, it would be removed. We used to periodically see things like what you’re showing, but starting with version 0.0.11 (released Sept 2021) it’s very unlikely the issue is within tedana. Can you check that your version is at least 0.0.11? (0.0.12 or pre-version release is recommended).
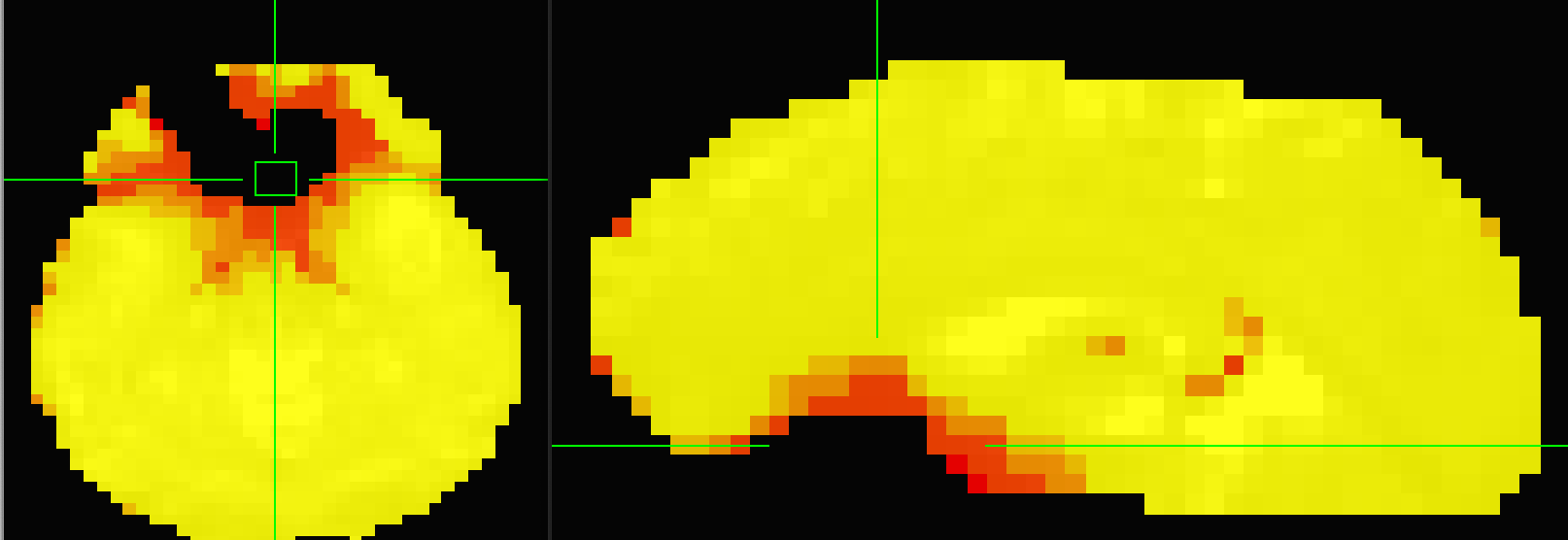
If you’re using default file names (and v0.0.11 or 12), there should be a file named desc-adaptiveGoodSignal_mask.nii.gz That is the mask tedana uses. You can look at it for some of your runs that do and don’t give reasonable results. The number is the number of echoes that tedana considers good. It should be all of your echoes for most of the brain, but will be lower in dropout regions. I’ve included a sample image for 5 echo data going from 5 (yellow) to 1 (red). Note that every voxel will be included in the final output, but only voxels with at least 3 echoes will be noised for denoising calculations.
Hello - yes we are using version 0.0.12. Judging by the fact that a) some of the people we see holes in their mask seem to have had acceptable motion artifact ranges (mean frame displacement below <=0.2) b) when using the --mask flag coupled with afni automask we see components picking up signal where there were holes before (meaning that there is BOLD signal there?) one could argue that the automatic masking is being a bit too stringent no?
We looked at the adaptive mask but because we only have 3 echoes all the masks always show the same thing for each echo.
ps.: the --mask+3dAutomask did help thank you! although now there is a question of whether for “3 echo datasets” is running tedana with --mask better than letting the denoising do its thing or not?
Thanks for your comments @dowdlelt and @handwerkerd. As my colleague Tamas says, the Automask worked (see below)
Inputting your own mask with the --mask option should still do tedana denoising and the results should be fine. Check your outputs (tedana_*.tsv) for any warnings that might be related to masking issues. Even with your own mask, adaptiveGoodSignal_mask.nii.gz should be generated and most of the values in the mask should be 3. If not, that would be a concern with the data (not necessarily tedana).
I don’t know why the automask is failing so spectacularly, but I’m glad a user-inputted mask is a successful alternative. I’m swamped with other things for the next several of weeks, but, if you can share a sample dataset where automask failed, I or another tedana developer might be able to take a look. (Prod me in a few weeks if there’s no reply)