Hello Neurostars community,
After doing preprocessing with fmriprep I have done some analysis in volume space [bold_space-T1w] and now I want to map these results (nii.gz) to surface (native or fsaverage) as an mgh file.
Freesurfer command that I think can do this [ mri_vol2surf] requires a source registration file [register.dat].
I know that there is this affine translation computed between T1w space and fsnative [sub_T1w_target-fsnative_affine.txt] with fmriprep.
Any ideas on how can I convert this transformation to the register.dat file that mri_vol2surf requires?
3 Likes
Hi @ilkay_isik. You may be able to use --regheader sub-01 instead of --reg <register.dat>. I would test this by using freeview to overlay freesurfer/sub-01/T1.mgz and your analysis results. If they appear in register, then it should be safe.
If not, then you can convert sub_T1w_target-fsnative_affine.txt to LTA (mri_vol2surf will accept LTAs instead of .dat files, and it’s their current recommendation) using lta_convert. I think there’s a way of testing your LTA file by applying it when loading your overlay image in freeview, but it’s been a bit since I played with it.
1 Like
Thanks so much @effigies. That was very helpful.
I tried both suggestions.
1st suggestion
I have used this command:
mri_vol2surf --src cope2.nii.gz --out lh.cope2.mgh \
--regheader sub-01 \
--hemi lh
This runs without an error and creates the output file but this file cannot be overlaid in freesurfer. I am not quite sure what exactly goes wrong here.
2nd suggestion
I used lta_convert as you suggested to convert the affine.txt and created both the .lta and the .dat files:
lta_convert --initk ${subj}_T1w_target-fsnative_affine.txt \
--outlta ${subj}_T1w_target-fsnative_affine.lta \
--outreg ${subj}_T1w_target-fsnative_affine.dat \
--src ${subj}_T1w_preproc.nii.gz \
--trg freesurfer/${subj}/mri/T1.mgz \
--subject ${subj}
When I want to use the .lta file as the registration file
mri_vol2surf --src cope2.nii.gz \
--out lh.cope2.mgh \
--srcreg ${subj}_T1w_target-fsnative_affine.lta \
--hemi $hemi
this gives an error : ERROR: source volume is neither source nor target of the registration
But when I use the .dat file as the registration file, it seems to work fine.
mri_vol2surf --src cope2.nii.gz \
--out lh.cope2.mgh \
--srcreg ${subj}_T1w_target-fsnative_affine.dat \
--hemi $hemi
I wonder why it does not work with lta.
[I use: freesurfer-Darwin-OSX-stable-pub-v6.0.0-2beb96c]
You might want to try using --src ${subj}_${task}_bold_preproc.nii.gz, so that the source image has the same geometry as your COPEs. It should be in the same space as T1w, so the RAS2RAS matrix shouldn’t change, just the source geometry.
Indeed your first suggestion ( using --regheader sub-01 instead of --reg <register.dat> using mri_vol2surf) seems to work completely fine.
I don’t know what went wrong last time.
I also compared the surface maps with my old pipeline results where I computed a registration between volume and surface using reg-feat2anat and the activations on the surface are strikingly similar.
Thanks so much again!
hmm, any ideas how to map these maps to fsaverage?
1 Like
Just add --trgsubject fsaverage. --srcsubject should be determined by --regheader, so no need to separately specify.
Did work again!
I guess another option would be using surf2surf from fsnative to fsaverage but I prefer your suggestion.
Thanks a lot.
1 Like
Any ideas on how would the reverse transform [from fsaverage to volume-MNI space] work? I am trying with surf2vol but does not work properly.
So fsaverage is in the FreeSurfer variant of the MNI305 space, which you could then potentially map to other MNI templates, but that would require a registration that I don’t know anybody has published.
Alternately, you can mri_vol2surf back into subject space and apply the ANTs transform (you might need to sync qform and sform to make sure ANTs applies it correctly). I’m not sure how much distortion you can expect to be added by the multiple interpolations.
But stepping back a bit, could you describe why you’re looking to move from one group space to another? There might be a cleaner solution that allows you to pick one group space to target and stay there (e.g. CIFTI-2 allows for side-by-side volumetric subcortical and surface-based cortical representations).
About this particular question (how to map from fsaverage to MNI) I was just curious.
But in general, I do going back and forth between surface and volume when defining the functional ROIs. I do the functional ROI creation process on the surface and then transform those ROIs back to volume and continue doing analysis in volume space. (For example, I would draw the ROI borders on fsnative surface [guided by the group results], then transform the label back to volume using label2vol.)
In my old pipeline [using reg-feat2anat to do volume to surface mapping] I would use the created registration file for many vol2surf or surf2vol operations.
Now that I am changing this pipeline I am trying to figure out how to do these transforms without having this reg file.
I see. At least for ROIs, I would say that it shouldn’t be too problematic to go through multiple interpolation steps using nearest-neighbor interpolation.
As for generating a registration file, you can probably use tkregister2 to create a file that is basically an identity transform that will work whenever you need something explicit, but as long as you use a file that’s in T1w as your reference, I think most FreeSurfer tools should just work, since the T1w space is in register* with the T1.mgz space.
To the best of my memory, it looks like you’re one of the first people really using FreeSurfer tools to work in both the FreeSurfer and fMRIPrep-generated spaces (as opposed to moving to a group space and stickig with it). Sorry that it’s not as straightforward as I might have hoped, but as you work through these, I think it would be a very valuable addition to our documentation to show how to go back and forth like this.
* With some caveats when you have multiple T1w images or you ran FreeSurfer prior to running fMRIPrep.
Until now with your help I have figured how to do the most important operations.
For example, I do a lot of mri_label2vol to carry roi’s that I draw on the surface back to volume. This requires anat2exf.register.dat in my old pipeline.
But looks like it also works fine when I use --identity as the reg method
[temp should be the orig.mgz]
mri_label2vol --subject sub-01 --hemi rh \
--identity \
--label path/ffa.label \
--temp path/freesurfer/sub-01/mri/orig.mgz \
--o path/ffa_label.nii
I could not figure out how to do the identity transform with tkregister2 but I tried bbregister:
bbregister --s sub-01 --mov fmriprep/sub-01/anat/sub-01_T1w_preproc.nii.gz \
--reg sub-01_register.dat --init-header --t1
which creates a registration file between t1w and orig.mgz.
and this reg file can then be used when doing the same mri_label2vol transformation [but this time the temp should be the T1w not orig.mgz]
mri_label2vol --subject sub-01 --hemi rh \
--label path/ffa.label \
--temp fmriprep/sub-01/anat/sub-01_T1w_preproc.nii.gz \
--o path/ffa_label2.nii
--reg sub-01_register.dat
I am still yet to figure out how to transform an .mgh file back to volume, but for now that is not crucial for my analysis.
I can gather this info together and create a documentation if you think that would be useful for others.
1 Like
Hello ilkay_isik and effigies,
Sorry in advance if this is a trivial question, I am very new to this and this is actually my first post!
I came across this topic because I am trying to do the exact same thing, mri_vol2surf using fmriprep outputs.
I did as suggested by effigies :
mri_vol2surf --src sub-CU0009_desc-preproc_T1w.nii.gz --out lh.sub-CU0009.mgh --regheader sub-CU0009 --hemi lh
And I have the output needed : lh.sub-CU0009.mgh
My question is, how do you visualize this .mgh file on freeview to see surfaces?
I would like to check if the command worked but I don’t understand how and wasn’t successful.
Thank you in advance!
Hi Idil,
You can load the left hemisphere surface and load the mgh file as an overlay.
You can do this via the gui of Freeview or using the command line with a code like this:
freeview -f ${SUBJECTS_DIR}/fsaverage/surf/lh.inflated:overlay=path/to/mgh/file.mgh
1 Like
Hi again !
So I used the recommended flags by @effigies and then overlayed as suggested by @ilkay_isik :
mri_vol2surf --src <participant_ID>_task-ert_space-T1w_desc-preproc_bold.nii.gz --out lh.<participant_ID>_ert_fsaverage_T1w.mgh --regheader <participant_ID> --hemi lh --trgsubject fsaverage
However, compared the a surface output processed with fmriprep, <participant_ID>_task-ert_space-fsaverage_hemi-L.func.gii , the output from fmriprep appears to have a smoother surface.
Do you know why this may be the case? Is it because of averaging signal along the cortical ribbon? It appears that from the Fmriprep documentation : “For each vertex, the cortical ribbon is sampled at six points (spaced 20% of thickness apart) and averaged." Is there a way to replicate this using freesurfer’s mri_vol2surf?
I tried to add the flag --projfrac-avg 1.000 as seen in the Nypipe fs.SampleToSurface used by fmriprep but it doesn’t seem to work in my implementation, I get an error.
To summarize my question: how can I make the output obtained from mri_vol2surf outside of the fmriprep pipeline more similar to the one from fmriprep?
Below are the outputs from fmriprep and mri_vol2surf :
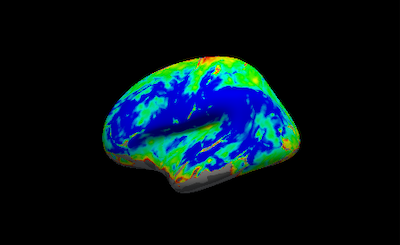
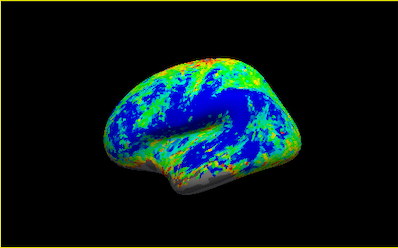
I think you want --projfrac-avg 0 1 0.2
Hi everyone! I tried out the 1st suggestion, but it looks like things may have changed between freesurfer versions (I am on 7.3.2). Here is my call to mri_vol2surf:
subid="MWMH212"
sesid="2"
ssfreedir="/projects/b1108/studies/mwmh/data/processed/neuroimaging/fmriprep_23.1.4/sourcedata/freesurfer/sub-"${subid}
ssprepdir="/projects/b1108/studies/mwmh/data/processed/neuroimaging/fmriprep_23.1.4/sub-"${subid}"/ses-"${sesid}
sssurfdir="/projects/b1108/studies/mwmh/data/processed/neuroimaging/surf/sub-"${subid}"/ses-"${sesid}
mri_vol2surf --src ${ssprepdir}/func/sub-${subid}_ses-${sesid}_task-rest_space-T1w_desc-preproc_bold.nii.gz \
--out ${sssurfdir}/sub-${subid}_ses-${sesid}_task-rest_space-T1w_desc-preproc_bold_lh.mgh \
--regheader sub-${subid} --hemi lh
When I try to run this, I get the following error: “error: mghRead(/hpc/software/freesurfer/7.3.2/freesurfer/subjects/sub-MWMH212/mri/orig.mgz, -1): could not open file”.
Clearly this file shouldn’t exist in this directory, but if I try to give the full path, I get a similar error:
mri_vol2surf --src ${ssprepdir}/func/sub-${subid}_ses-${sesid}_task-rest_space-T1w_desc-preproc_bold.nii.gz \
--out ${sssurfdir}/sub-${subid}_ses-${sesid}_task-rest_space-T1w_desc-preproc_bold_lh.mgh \
--regheader ${ssfreedir} --hemi lh
“error: mghRead(/hpc/software/freesurfer/7.3.2/freesurfer/subjects//projects/b1108/studies/mwmh/data/processed/neuroimaging/fmriprep_23.1.4/sourcedata/freesurfer/sub-MWMH212/mri/orig.mgz, -1): could not open file”
Is there a way to specify the full path to the orig.mgz file? If not, is there another flag I could use? This command works if I change --regheader to fsaverage5 and set the src image to be in MNI space, but I think it would be better to go through anat space to the subject’s surface space to fsaverage5 so as to avoid data loss.
Answered my question using ChatGPT! I had to set SUBJECTS_DIR="/projects/b1108/studies/mwmh/data/processed/neuroimaging/fmriprep_23.1.4/sourcedata/freesurfer"
3 Likes

