Hi, I’m doing my intern related to CT images. Now I was very confused about voxel depth and CT image thickness. For example, I am checking a CT scan(NIFTI format), I nibabel.header.get_zooms() to get the voxel size and it was 0.6X0.6X1, which means the voxel depth is 1. However, the thickness is 2. So Does it merge two voxels from the z-line into one? If it is in that case, would it be possible that when they construct the slice there are overlap between voxels? Like, First slice = Voxels[:,:,0:2],(x,y,z)
Second slice = Voxels[:,:,1:3]… Thanks in advance if anyone can help!!
What tool was used to convert your DICOM images to NIfTI format. If you used dcm2niix, you may be interested in the notes here as well as the validation dataset here. The NIfTI format was initially created for MR data, where slices for a series are always equidistant and the q-form assumes that volumes do not show shear (the s-form can encode shear, but many tools ignore that).
In contrast, CT scans often have close slices at the critical brain stem with a wider gap between slices covering the cortex. Further, CT scans are often acquired with gantry tilt, that yields volumes with shear. When converting scans, dcm2niix detects these differences and interpolates slices. You can look at the dcm2niix warnings when it converts images to see if these manipulations are required. If your volume is not rhomboidal and the slices are equidistant, the output NIfTI will match the input DICOM.
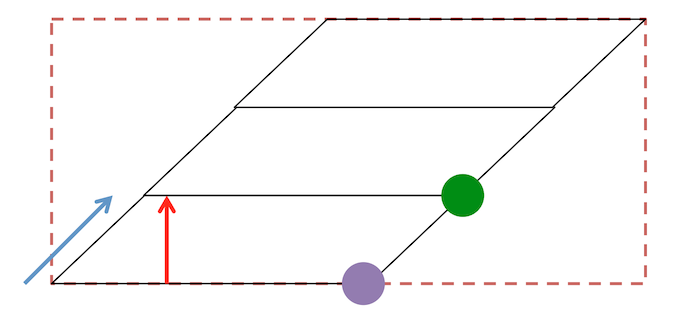
Also be aware that the NIfTI pixdim[3] reports the distance between voxel slice centers: in other words this reflects the combination of the slice gap and the slice thickness. Therefore, if a series uses a 1.2mm slice thickness, with a 0.8mm slice gap, the pixdim[3] is 2.0mm.