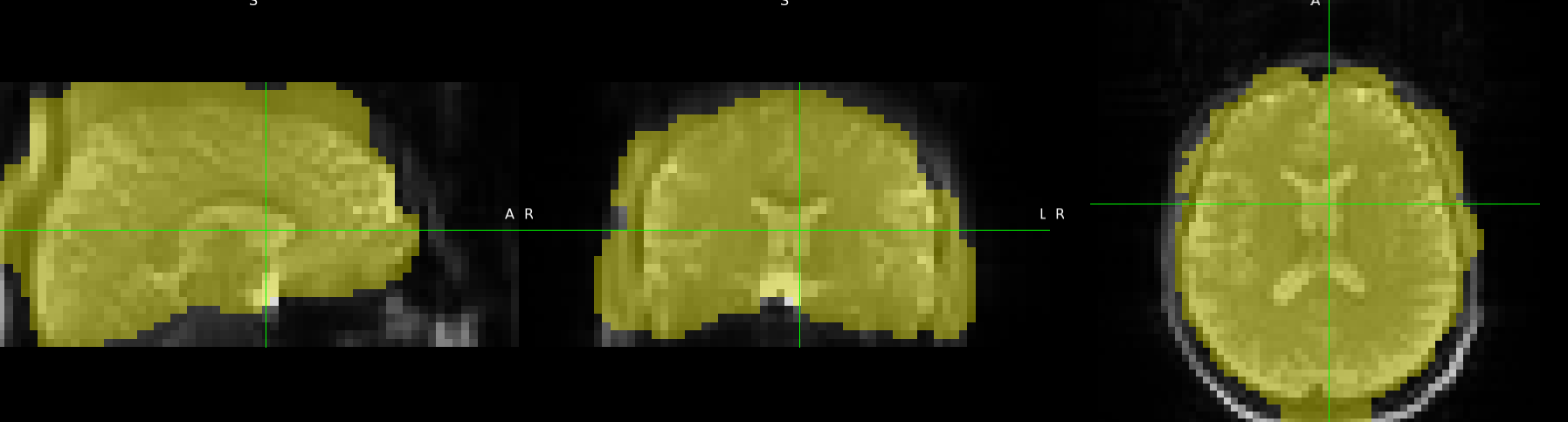
Summary of what happened:
I tried running ASLPrep on several subjects, but the brain masks it produced were all very strange. In contrast, when I applied BET directly, the results were correct.
Command used (and if a helper script was used, a link to the helper script or the command generated):
BIDS=/gs/gsfs0/users/BIDS/HP
OUT=/gs/gsfs0/users/ASL_out
WORK=/gs/gsfs0/users/ASL_work
FS_DIR=/gs/gsfs0/users/BIDS/freesurfer
FS_LICENSE=/gs/gsfs0/users/BIDS/freesurfer/license.txt
IMG=~/toolbox/aslprep/aslprep-25.1.0.sif
SUB=sub-003
mkdir -p "$OUT" "$WORK"
singularity run --cleanenv \
-B "$BIDS":/bids:ro \
-B "$OUT":/out \
-B "$WORK":/work \
-B "$FS_DIR":/fsdir \
-B "$FS_LICENSE":/license.txt \
"$IMG" \
/bids /out participant \
--participant-label "$SUB" \
--nprocs 8 \
--omp-nthreads 8 \
--mem 32GB \
--basil \
--output-spaces T1w \
--fs-license-file /license.txt \
--fs-subjects-dir ${FS_DIR} \
--fs-no-reconall
Version:
25.1.0
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
My first asl volume is M0, but second is a dummy scan. I just set the seconde line as noRF
volume_type
m0scan
noRF
label
control
label
control
label
control
label
...
Here is my json file for ASL
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 123.248,
"Manufacturer": "Siemens",
"ManufacturersModelName": "Prisma",
"BodyPartExamined": "HEAD",
"PatientPosition": "HFS",
"SoftwareVersions": "syngo MR E11",
"MRAcquisitionType": "3D",
"SeriesDescription": "tgse_pcasl_Multi_Delay_ve11e_3.5iso_default",
"ProtocolName": "tgse_pcasl_Multi_Delay_ve11e_3.5iso_default",
"ScanningSequence": "EP",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"SequenceName": "tgse3d1_2268",
"ImageType": ["ORIGINAL", "PRIMARY", "ASL", "NONE", "ND", "MOSAIC"],
"NonlinearGradientCorrection": false,
"SeriesNumber": 10,
"AcquisitionTime": "15:47:2.145000",
"AcquisitionNumber": 1,
"SliceThickness": 3.5,
"SpacingBetweenSlices": 3.5,
"SAR": 0.103204,
"EchoTime": 0.03168,
"RepetitionTime": 4.35,
"FlipAngle": 120,
"RFGap": 0.00036,
"MeanGzx10": 6,
"ArterialSpinLabelingType": "PCASL",
"M0Type": "Included",
"RepetitionTimePreparation": 4.35,
"LabelingDuration": 0.7,
"PostLabelingDelay": 1.8,
"BackgroundSuppression": true,
"LabelingEfficiency": 0.85,
"ArterialSpinLabelingType": "PCASL",
"AcquisitionVoxelSize": [
3.5,
3.5,
3.5 ],
"PartialFourier": 1,
"BaseResolution": 64,
"ShimSetting": [
4321,
-1983,
5074,
576,
72,
187,
14,
35 ],
"TxRefAmp": 216.302,
"PhaseResolution": 0.984375,
"ReceiveCoilName": "HeadNeck_64",
"ReceiveCoilActiveElements": "HC3-6",
"PulseSequenceDetails": "%CustomerSeq%\\tgse_pcasl_ve11e",
"CoilCombinationMethod": "Sum of Squares",
"ConsistencyInfo": "N4_VE11E_LATEST_20181129",
"MatrixCoilMode": "SENSE",
"RepetitionTimePreparation": 4350,
"PercentPhaseFOV": 100,
"PercentSampling": 98.4375,
"EchoTrainLength": 63,
"PhaseEncodingSteps": 63,
"AcquisitionMatrixPE": 63,
"ReconMatrixPE": 64,
"PixelBandwidth": 3125,
"DwellTime": 2.5e-06,
"PhaseEncodingDirection": "j-",
"ImageOrientationPatientDICOM": [
1,
0,
0,
0,
0.999781,
-0.0209424 ],
"ImageOrientationText": "Tra>Cor(-1.2)",
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20220720"
}