Summary of what happened:
Dear all,
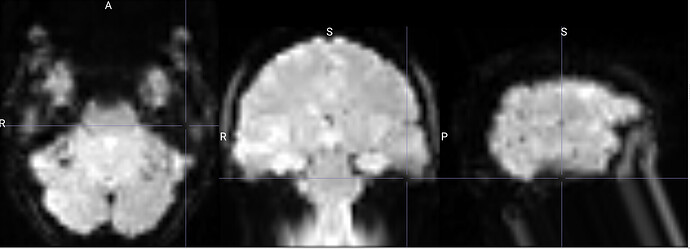
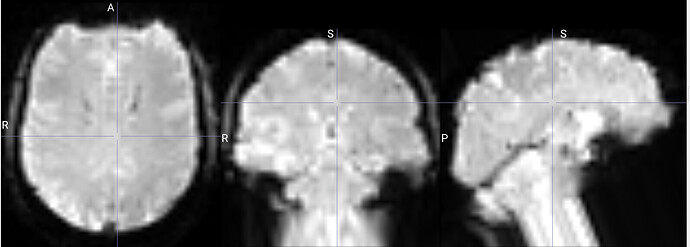
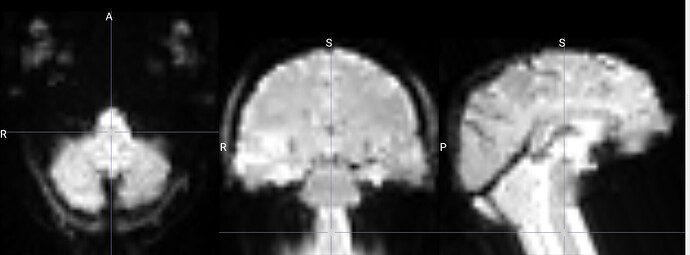
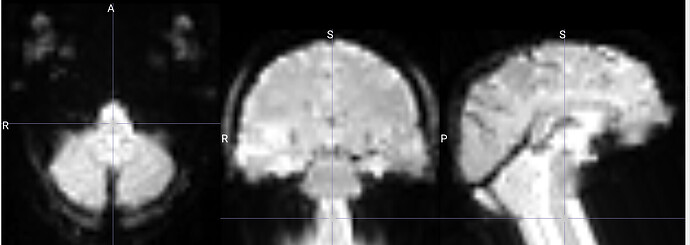
I am running fmriprep for the first time, and my preproc_bold.nii files have a weird distortion extending from the cerebellum for a few slices, it is basically a copy of what seems like the final slice, extending anteriorly.
The copied slices are overlapping with the superior slices, likely messing with the voxels.
I am currently re-running without --fs-no-reconall \ to see how it acts with freesurfer.
* Original orientation: LAS
* Repetition time (TR): 2s
* Phase-encoding (PE) direction: Anterior-Posterior
* Single-echo EPI sequence.
* Slice timing correction: Applied
* Susceptibility distortion correction: None
* Registration: FSL `flirt` with boundary-based registration (BBR) metric - 6 dof
* Non-steady-state volumes: 0
Also I must note that this issue does not happen at my own SPM preprocessing pipeline.
images showing how it extends anteriorly:
Command used (and if a helper script was used, a link to the helper script or the command generated):
my fmriprep command is:
fmriprep /neurodesktop-storage/analysis/nifti /neurodesktop-storage/analysis/derivatives participant \
--work-dir /neurodesktop-storage/analysis/work \
--fs-license-file /neurodesktop-storage/analysis/license.txt \
--output-spaces T1w MNI152NLin2009cAsym \
--nthreads 12 \
--omp-nthreads 6 \
--mem-mb 16000 \
--fs-no-reconall \
--skip-bids-validation
Version:
25.1.3
Environment (Docker, Singularity / Apptainer, custom installation):
Neurodesk
Data formatted according to a validatable standard? Please provide the output of the validator:
It is BIDS validated, no major errors, only irrelevant warnings.