Dear Neurostars,
we are trying to preprocess our DWI data using qsiprep, which has been great so far. We do however run into an issue with the SDC.
Any help would be appriciated!
Thank you so much!
Summary of what happened:
After qsiprep is applied to our DWI (to runs, one with 98 dirs, one with 99 dirs, both AP), we have strange “shadows” anterior and posterior (see attached gif). These seem to appear during SDC (using TOPUP, with an APPA scheme). To us, this does not look like it is intended. Do you have any guidance / ideas, what is going wrong?
Command used (and if a helper script was used, a link to the helper script or the command generated):
command = [
"docker", "run", "--rm",
"-v", f"{bids_dir}:/data:ro",
"-v", f"{output_dir}:/out",
"-v", f"{work_dir}:/work",
"pennlinc/qsiprep:1.0.1",
"/data", "/out", "participant",
"--work-dir", "/work",
"--bids-filter-file", "/data/code/BIDS_filter_file.json",
"--nprocs", "8",
"--mem", "16",
"--participant-label", bids_id,
"--unringing-method", "mrdegibbs",
"--denoise-after-combining",
"--output-resolution", "1.5",
"--skip-bids-validation"
]
Version:
1.0.1
Environment (Docker, Singularity / Apptainer, custom installation):
We are using docker.
Data formatted according to a validatable standard? Please provide the output of the validator:
└── ses-01
├── anat
│ ├── sub-31_ses-01_T1w.json
│ └── sub-31_ses-01_T1w.nii.gz
├── dwi
│ ├── sub-31_ses-01_dir-AP_run-01_sbref.json
│ ├── sub-31_ses-01_dir-AP_run-01_sbref.nii.gz
│ ├── sub-31_ses-01_dir-AP_run-02_sbref.json
│ ├── sub-31_ses-01_dir-AP_run-02_sbref.nii.gz
│ ├── sub-31_ses-01_run-01_dwi.bval
│ ├── sub-31_ses-01_run-01_dwi.bvec
│ ├── sub-31_ses-01_run-01_dwi.json
│ ├── sub-31_ses-01_run-01_dwi.nii.gz
│ ├── sub-31_ses-01_run-02_dwi.bval
│ ├── sub-31_ses-01_run-02_dwi.bvec
│ ├── sub-31_ses-01_run-02_dwi.json
│ └── sub-31_ses-01_run-02_dwi.nii.gz
└── fmap
├── sub-31_ses-01_dir-AP_epi.json
├── sub-31_ses-01_dir-AP_epi.nii.gz
├── sub-31_ses-01_dir-PA_epi.json
└── sub-31_ses-01_dir-PA_epi.nii.gz
Relevant log outputs (up to 20 lines):
Summary
Subject ID: sub-31
Structural images: 1 T1-weighted
Diffusion-weighted series: inputs 4, outputs 1
Output Name: sub-31_ses-01
Scan group: sub-31_ses-01 (PE Dir j-)
DWI Files:
/data/sub-31/ses-01/dwi/sub-31_ses-01_run-01_dwi.nii.gz
/data/sub-31/ses-01/dwi/sub-31_ses-01_run-02_dwi.nii.gz
Fieldmap type: epi
suffix: epi
epi: [‘/data/sub-31/ses-01/fmap/sub-31_ses-01_dir-AP_epi.nii.gz’, ‘/data/sub-31/ses-01/fmap/sub-31_ses-01_dir-PA_epi.nii.gz’]
Resampling targets: ACPC
Transform targets: [‘ACPC’, ‘MNI152NLin2009cAsym’]B0 field mapping
Reports for: session 01.
A total of 2 distortion groups were included in the data data. Distortion group ‘0 -1 0 0.095910’ was represented by image 1 from sub-31_ses-01_run-02_dwi.nii.gz. Distortion group ‘0 1 0 0.095910’ was represented by image 0 from sub-31_ses-01_dir-PA_epi.nii.gz.Reports for: session 01.
Summary
Phase-encoding (PE) direction: Anterior-Posterior
Susceptibility distortion correction: TOPUP
Screenshots / relevant information:
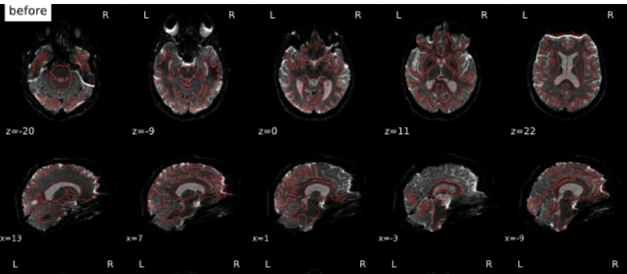
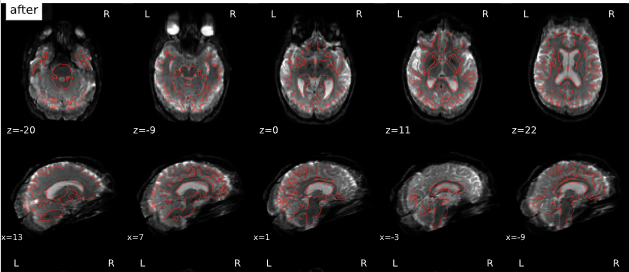
Not sure whether the upload of the images worked.