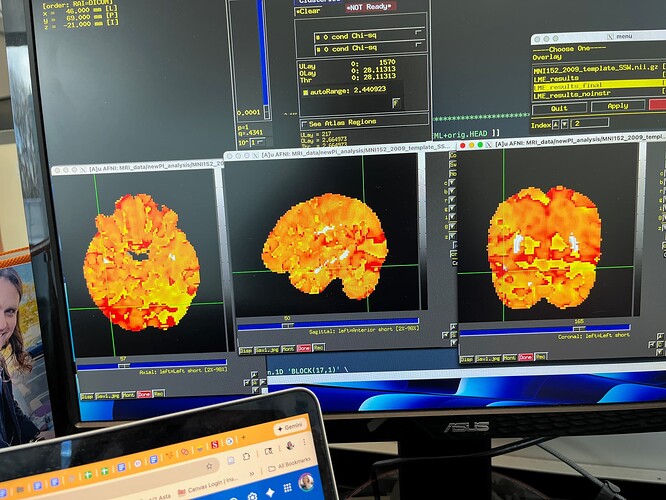
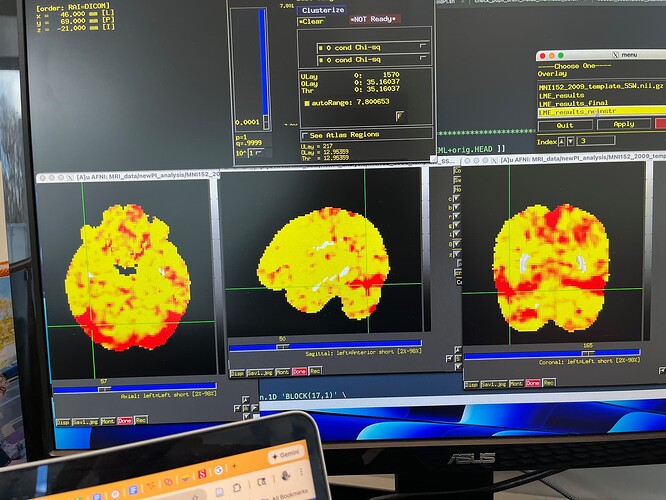
Summary of what happened: I made an afni_proc.py pipeline, and the final second level results looked really, really strange.
(See picture below for an example. Please note that only the Chi-squared tests have this glitchy look. The contrasts I specified do not look glitchy.) Because I needed to save money on compute costs, I extracted the part of the pipeline beginning at the regress block (everything before that looked great), and redid it, with a few changes to 3dDeconvolve (removed motion regression since I’m using multi-echo data anyway). Originally, to test whether those changes were successful, I only redid 3dDeconvolve and 3dREMLfit. This pipeline does not include alignment to standard space, but I calculated warps with SSWarper2 earlier and used those warps to align each person’s individual first level stats bucket to MNI space. I then ran 3dLMEr for second level analysis. The results looked great! The pattern of results was very similar to the glitchy version (in terms of the specific contrasts; the Chi-squared tests looked normal and correct, which bore no resemblance to the glitchy first version), but the p values were about 0.1-0.2 lower across the board, creating a completely reasonable pattern of results that made sense. Thinking I had successfully diagnosed my pipeline issue, and wanting the entire pipeline to be consistent/complete (especially the TSNR calculations), I replaced my newly created stats outputs with a “final” version, which still wasn’t the full pipeline from start to finish, but which took the entire afni_proc.py script starting at the regress block, translated it to bash, and ran it to the very end through QC. The output of that version looks just as terrible as it did the first time (to be clear, in exactly the same glitchy way). I’ve spent an embarrassing number of hours reviewing everything and can’t find the problem. Running the entire pipeline start to finish takes around 2 weeks and is quite costly, and I can’t afford to just try it all over again without any inkling of what caused this.
In the code below, you will note I have commented out blur calculations since it’s so slow.
Things that have not changed:
-these versions use exactly the same input files to 3dDeconvolve; they were not recalculated
-the timing files have not changed
-the 3dLMEr code for second level analysis has not changed
-the warps to MNI space have not changed
-the first level individual deconvolutions looked normal in both cases and did not have this patchy/glitchy look at all
Another important fact: I copy-pasted this adapted end-of-pipeline (the code below) and redid the end of a different pipeline also. The results of that one look great (not weird). Same exact code except for file paths and the 3dDeconvolve block.
Command used (and if a helper script was used, a link to the helper script or the command generated):
#!/bin/bash
#SBATCH -n 4
#SBATCH -N 4
#SBATCH -c 4
#SBATCH -J PI_native_decon
#SBATCH -o sbatch-%j.out
#SBATCH -e sbatch-%j.err
#SBATCH --mail-type=ALL
#SBATCH --oversubscribe
#SBATCH --ntasks-per-core=1
#SBATCH -exclude=aclab-cluster-c-0
module load afni
# directories that will de-clutter code below
base_folder=/home/shared/aclab-fmri/Studies/33_MOTIP2018
preproc_results=${base_folder}/MRI_data/PREPROCESSED/results
old_subject_folder=${base_folder}/MRI_data/locations/PROI2_subjects.txt
subject_folder=${base_folder}/MRI_data/locations/newPI2_subjects.txt
old_subjects=`cat ${old_subject_folder}`
subjects=`cat ${subject_folder} `
#loop through all subjects that you input above
for subject in 510 # $subjects
do
subj=${subject: -3}
echo $subj
echo 'Beginning ******************************************************************************************'
if [[ ! -f ${preproc_results}/${subj}_newPI_native.results/stats.${subj}_REML+orig.HEAD ]]
then
echo "Deconvolution for ${subj} was completed already, but we are removing and doing again"
rm ${preproc_results}/${subj}_newPI_native.results/stats.REML_cmd
rm ${preproc_results}/${subj}_newPI_native.results/stats.${subj}_REML+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/stats.${subj}_REMLvar+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/stats.${subj}+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/stats.${subj}_REML+tlrc.*
rm ${preproc_results}/${subj}_newPI_native.results/stats.${subj}+tlrc.*
rm ${preproc_results}/${subj}_newPI_native.results/errts.${subj}_REML+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/errts.${subj}+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/fitts.${subj}_REML+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/fitts.${subj}+orig.*
rm ${preproc_results}/${subj}_newPI_native.results/TSNR.${subj}*
#else
#echo "Deconvolution for ${subj} is beginning"
cd ${preproc_results}/${subj}_newPI_native.results
{
echo $subj
# ------------------------------ pasting this in from the end of the original afni_proc script, just changing the 3dDeconvolve part
# run the regression analysis
3dDeconvolve -input pb06.${subj}.r*.scale+orig.HEAD \
-censor censor_${subj}_combined_2.1D \
-num_stimts 10 \
-polort A \
-stim_times 1 stimuli/r1_h1_on.1D 'BLOCK(17,1)' \
-stim_label 1 BASE_H1 \
-stim_times 2 stimuli/r1_h2_on.1D 'BLOCK(17,1)' \
-stim_label 2 BASE_H2 \
-stim_times 3 stimuli/PI_q1_on.1D 'BLOCK(17,1)' \
-stim_label 3 PI_Q1 \
-stim_times 4 stimuli/PI_q2_on.1D 'BLOCK(17,1)' \
-stim_label 4 PI_Q2 \
-stim_times 5 stimuli/PI_q3_on.1D 'BLOCK(17,1)' \
-stim_label 5 PI_Q3 \
-stim_times 6 stimuli/PI_q4_on.1D 'BLOCK(17,1)' \
-stim_label 6 PI_Q4 \
-stim_times 7 stimuli/NL_q1_on.1D 'BLOCK(17,1)' \
-stim_label 7 NL_Q1 \
-stim_times 8 stimuli/NL_q2_on.1D 'BLOCK(17,1)' \
-stim_label 8 NL_Q2 \
-stim_times 9 stimuli/NL_q3_on.1D 'BLOCK(17,1)' \
-stim_label 9 NL_Q3 \
-stim_times 10 stimuli/NL_q4_on.1D 'BLOCK(17,1)' \
-stim_label 10 NL_Q4 \
-jobs 8 \
-gltsym 'SYM: +BASE_H1 +BASE_H2 +PI_Q1 +PI_Q2 +PI_Q3 +PI_Q4 +NL_Q1 +NL_Q2 +NL_Q3 +NL_Q4' \
-glt_label 1 ONvOFF \
-gltsym 'SYM: +0.5*BASE_H1 +0.5*BASE_H2 -0.25*PI_Q1 -0.25*PI_Q2 -0.25*PI_Q3 -0.25*PI_Q4' \
-glt_label 2 BASEvsPI \
-gltsym 'SYM: +0.5*BASE_H1 +0.5*BASE_H2 -0.25*NL_Q1 -0.25*NL_Q2 -0.25*NL_Q3 -0.25*NL_Q4' \
-glt_label 3 BASEvsNL \
-gltsym 'SYM: +BASE_H2 -PI_Q1' \
-glt_label 4 BASEvsPI_specific \
-gltsym 'SYM: +BASE_H2 -NL_Q1' \
-glt_label 5 BASEvsNL_specific \
-gltsym 'SYM: +PI_Q1 +PI_Q2 +PI_Q3 +PI_Q4 -NL_Q1 -NL_Q2 -NL_Q3 -NL_Q4' \
-glt_label 6 PIvsNL \
-gltsym 'SYM: +PI_Q1 -NL_Q1' \
-glt_label 7 PIvsNL_specific \
-gltsym 'SYM: +PI_Q1 -0.33*PI_Q2 -0.33*PI_Q3 -0.33*PI_Q4' \
-glt_label 8 PI1vsPI234 \
-gltsym 'SYM: +NL_Q1 -0.33*NL_Q2 -0.33*NL_Q3 -0.33*NL_Q4' \
-glt_label 9 NL1vsNL234 \
-fout -tout -x1D X.xmat.1D -xjpeg X.jpg \
-x1D_uncensored X.nocensor.xmat.1D \
-fitts fitts.${subj} \
-errts errts.${subj} \
-bucket stats.${subj}
# display any large pairwise correlations from the X-matrix
1d_tool.py -show_cormat_warnings -infile X.xmat.1D 2>&1 | tee out.cormat_warn.txt
# display degrees of freedom info from X-matrix
1d_tool.py -show_df_info -infile X.xmat.1D 2>&1 | tee out.df_info.txt
# -- execute the 3dREMLfit script, written by 3dDeconvolve --
tcsh -x stats.REML_cmd
# create an all_runs dataset to match the fitts, errts, etc.
3dTcat -prefix all_runs.${subj} pb06.${subj}.r*.scale+orig.HEAD
# --------------------------------------------------
# create a temporal signal to noise ratio dataset
# signal: if 'scale' block, mean should be 100
# noise : compute standard deviation of errts
ktrs=$(1dcat out.keep_trs_rall.txt | tr ' ' ',')
3dTstat -mean -prefix rm.signal.all "all_runs.${subj}+orig[${ktrs}]"
3dTstat -stdev -prefix rm.noise.all "errts.${subj}_REML+orig[${ktrs}]"
3dcalc -a rm.signal.all+orig \
-b rm.noise.all+orig \
-expr 'a/b' -prefix TSNR.${subj}
# --------------------------------------------------
# compute TSNR stats for dset labels: brain
compute_ROI_stats.tcsh \
-out_dir tsnr_stats_regress \
-stats_file tsnr_stats_regress/stats_auto_brain.txt \
-dset_ROI mask_epi_anat.${subj}+orig \
-dset_data TSNR.${subj}+orig \
-rset_label brain \
-rval_list 1
# ---------------------------------------------------
# compute and store GCOR (global correlation average)
# (sum of squares of global mean of unit errts)
3dTnorm -norm2 -prefix rm.errts.unit errts.${subj}_REML+orig
3dmaskave -quiet -mask mask_epi_anat.${subj}+orig \
rm.errts.unit+orig > mean.errts.unit.1D
3dTstat -sos -prefix - mean.errts.unit.1D\' > out.gcor.1D
echo "-- GCOR = $(cat out.gcor.1D)"
# ---------------------------------------------------
# compute correlation volume
# (per voxel: correlation with masked brain average)
3dmaskave -quiet -mask mask_epi_anat.${subj}+orig \
errts.${subj}_REML+orig > mean.errts.1D
3dTcorr1D -prefix corr_brain errts.${subj}_REML+orig mean.errts.1D
# create ideal files for fixed response stim types
1dcat X.nocensor.xmat.1D'[8]' > ideal_BASE_H1.1D
1dcat X.nocensor.xmat.1D'[9]' > ideal_BASE_H2.1D
1dcat X.nocensor.xmat.1D'[10]' > ideal_PI_Q1.1D
1dcat X.nocensor.xmat.1D'[11]' > ideal_PI_Q2.1D
1dcat X.nocensor.xmat.1D'[12]' > ideal_PI_Q3.1D
1dcat X.nocensor.xmat.1D'[13]' > ideal_PI_Q4.1D
1dcat X.nocensor.xmat.1D'[14]' > ideal_NL_Q1.1D
1dcat X.nocensor.xmat.1D'[15]' > ideal_NL_Q2.1D
1dcat X.nocensor.xmat.1D'[16]' > ideal_NL_Q3.1D
1dcat X.nocensor.xmat.1D'[17]' > ideal_NL_Q4.1D
# --------------------------------------------------
# extract non-baseline regressors from the X-matrix,
# then compute their sum
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
# # ============================ blur estimation =============================
# # compute blur estimates
# touch blur_est.${subj}.1D # start with empty file
# # create directory for ACF curve files
# mkdir -p files_ACF
# # -- estimate blur for each run in epits --
# touch blur.epits.1D
# # restrict to uncensored TRs, per run
# for run in 01 02; do
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# if [[ -z "$trs" ]]; then continue; fi
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.epits.r${run}.1D \
# "all_runs.${subj}+orig[${trs}]" >> blur.epits.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\')
# echo "average epits FWHM blurs: $blurs"
# echo "$blurs # epits FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\')
# echo "average epits ACF blurs: $blurs"
# echo "$blurs # epits ACF blur estimates" >> blur_est.${subj}.1D
# # -- estimate blur for each run in errts --
# touch blur.errts.1D
# for run in 01 02; do
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# if [[ -z "$trs" ]]; then continue; fi
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.errts.r${run}.1D \
# "errts.${subj}+orig[${trs}]" >> blur.errts.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\')
# echo "average errts FWHM blurs: $blurs"
# echo "$blurs # errts FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\')
# echo "average errts ACF blurs: $blurs"
# echo "$blurs # errts ACF blur estimates" >> blur_est.${subj}.1D
# # -- estimate blur for each run in err_reml --
# touch blur.err_reml.1D
# for run in 01 02; do
# echo "processing run $run"
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# if [[ -z "$trs" ]]; then continue; fi
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r${run}.1D \
# "errts.${subj}_REML+orig[${trs}]" >> blur.err_reml.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\')
# echo "average err_reml FWHM blurs: $blurs"
# echo "$blurs # err_reml FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\')
# echo "average err_reml ACF blurs: $blurs"
# echo "$blurs # err_reml ACF blur estimates" >> blur_est.${subj}.1D
# # add 3dClustSim results as attributes to any stats dset
# mkdir -p files_ClustSim
# # Get the last ACF line and the 3 lines before it
# acf_comment_line=$(grep -n "err_reml ACF blur estimates" blur_est.${subj}.1D | tail -1 | cut -d: -f1)
# if [[ -n "$acf_comment_line" ]]; then
# # Get the 4 lines ending with that comment (the comment line and 3 lines before it)
# acf_data=$(sed -n "$((acf_comment_line-3)),$acf_comment_line p" blur_est.${subj}.1D)
# echo "ACF data block:"
# echo "$acf_data"
# # Extract just the numbers from those 4 lines, take the first 3
# params=($(echo "$acf_data" | grep -oE '[0-9]+\.[0-9]+' | head -3))
# echo "Extracted params: '${params[0]}' '${params[1]}' '${params[2]}'"
# # Verify we have 3 parameters and first is between 0 and 1
# if [[ ${#params[@]} -eq 3 ]] && (( $(echo "${params[0]} > 0 && ${params[0]} < 1" | bc -l) )); then
# echo "ACF parameters are valid: a=${params[0]}, b=${params[1]}, c=${params[2]}"
# 3dClustSim -both -mask mask_epi_anat.${subj}+orig -acf ${params[0]} ${params[1]} ${params[2]} \
# -cmd 3dClustSim.ACF.cmd -prefix files_ClustSim/ClustSim.ACF
# else
# echo "ERROR: Invalid ACF parameters"
# exit 1
# fi
# else
# echo "ERROR: Could not find err_reml ACF blur estimates line"
# exit 1
# fi
# # Fix the 3drefit command
# if [[ -f 3dClustSim.ACF.cmd ]]; then
# echo "Running 3drefit command:"
# cat 3dClustSim.ACF.cmd
# # Execute the command from the file, adding the dataset names
# cmd_line=$(cat 3dClustSim.ACF.cmd)
# $cmd_line stats.${subj}+orig stats.${subj}_REML+orig
# else
# echo "3dClustSim.ACF.cmd not found, skipping 3drefit step"
# fi
# ========================= auto block: QC_review ==========================
# generate quality control review scripts and HTML report
# generate a review script for the unprocessed EPI data
# (all echoes of all runs)
gen_epi_review.py -script @epi_review.${subj} \
-dsets pb00.${subj}.r*.e*.tcat+orig.HEAD
# -------------------------------------------------
# generate scripts to review single subject results
# (try with defaults, but do not allow bad exit status)
# write AP uvars into a simple txt file
cat << EOF > out.ap_uvars.txt
mot_limit : 0.5
out_limit : 0.05
copy_anat : MPRAGE+orig.HEAD
combine_method : m_tedana
mask_dset : mask_epi_anat.${subj}+orig.HEAD
ss_review_dset : out.ss_review.${subj}.txt
echo_times : 14.0 27.0 40.0
max_4095_warn_dset : out.4095_warn.txt
reg_echo : 2
vlines_tcat_dir : vlines.pb00.tcat
EOF
# and convert the txt format to JSON
cat out.ap_uvars.txt | afni_python_wrapper.py -eval "data_file_to_json()" \
> out.ap_uvars.json
# initialize gen_ss_review_scripts.py with out.ap_uvars.json
gen_ss_review_scripts.py -exit0 \
-init_uvars_json out.ap_uvars.json \
-write_uvars_json out.ss_review_uvars.json
# ========================== auto block: finalize ==========================
# remove temporary files
rm -f rm.*
# --------------------------------------------------
# if the basic subject review script is here, run it
# (want this to be the last text output)
if [[ -e @ss_review_basic ]]; then
./@ss_review_basic 2>&1 | tee out.ss_review.${subj}.txt
# generate html ss review pages
# (akin to static images from running @ss_review_driver)
apqc_make_tcsh.py -review_style pythonic -subj_dir . \
-uvar_json out.ss_review_uvars.json
apqc_make_html.py -qc_dir QC_${subj}
echo -e "\nconsider running: \n"
echo " afni_open -b ${subj}_newPI_native.results/QC_${subj}/index.html"
echo ""
fi
} 2>&1 | tee "/home/shared/aclab-fmri/Studies/33_MOTIP2018/MRI_data/PREPROCESSED/results/output.proc.${subj}_newPI_native"
# return to parent directory (just in case...)
cd ${preproc_results}
echo "execution finished: `date`"
fi
done
# AND NOW FOR THE OLD SUBJECTS %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
# for subject in $old_subjects
# do
# subj=${subject: -3}
# echo $subj
# if [[ -f ${preproc_results}/${subj}_oldPI_native.results/stats.${subj}_REML+orig.HEAD ]]
# then
# echo "Deconvolution for ${subj} was completed already"
# else
# cd ${preproc_results}/${subj}_oldPI_native.results
# echo 'Beginning ******************************************************************************************'
# {
# echo $subj
# # ------------------------------
# # run the regression analysis
# 3dDeconvolve -input pb06.${subj}.r*.scale+orig.HEAD \
# -censor censor_${subj}_combined_2.1D \
# -polort A \
# -num_stimts 6 \
# -stim_times 1 stimuli/r1_h1_on.1D 'BLOCK(17,1)' \
# -stim_label 1 ON_H1_R1 \
# -stim_times 2 stimuli/r1_h2_on.1D 'BLOCK(17,1)' \
# -stim_label 2 ON_H2_R1 \
# -stim_times 3 stimuli/r2_q1_on.1D 'BLOCK(17,1)' \
# -stim_label 3 ON_Q1_R2 \
# -stim_times 4 stimuli/r2_q2_on.1D 'BLOCK(17,1)' \
# -stim_label 4 ON_Q2_R2 \
# -stim_times 5 stimuli/r2_q3_on.1D 'BLOCK(17,1)' \
# -stim_label 5 ON_Q3_R2 \
# -stim_times 6 stimuli/r2_q4_on.1D 'BLOCK(17,1)' \
# -stim_label 6 ON_Q4_R2 \
# -jobs 8 \
# -gltsym 'SYM: +ON_H1_R1 +ON_H2_R1 +ON_Q1_R2 +ON_Q2_R2 +ON_Q3_R2 +ON_Q4_R2' \
# -glt_label 1 ONvOFF \
# -gltsym 'SYM: +0.5*ON_H1_R1 +0.5*ON_H2_R1 -0.25*ON_Q1_R2 -0.25*ON_Q2_R2 -0.25*ON_Q3_R2 -0.25*ON_Q4_R2' \
# -glt_label 2 BASEvsPI \
# -gltsym 'SYM: +ON_H2_R1 -ON_Q1_R2' \
# -glt_label 3 BASEvsPI_specific \
# -gltsym 'SYM: +ON_Q1_R2 -0.33*ON_Q2_R2 -0.33*ON_Q3_R2 -0.33*ON_Q4_R2' \
# -glt_label 4 PI1vsPI234 \
# -fout -tout -x1D X.xmat.1D -xjpeg X.jpg \
# -x1D_uncensored X.nocensor.xmat.1D \
# -fitts fitts.${subj} \
# -errts errts.${subj} \
# -bucket stats.${subj}
# # display any large pairwise correlations from the X-matrix
# 1d_tool.py -show_cormat_warnings -infile X.xmat.1D 2>&1 | tee out.cormat_warn.txt
# # display degrees of freedom info from X-matrix
# 1d_tool.py -show_df_info -infile X.xmat.1D 2>&1 | tee out.df_info.txt
# # -- execute the 3dREMLfit script, written by 3dDeconvolve --
# tcsh -x stats.REML_cmd
# # create an all_runs dataset to match the fitts, errts, etc.
# 3dTcat -prefix all_runs.${subj} pb06.${subj}.r*.scale+orig.HEAD
# # --------------------------------------------------
# # create a temporal signal to noise ratio dataset
# # signal: if 'scale' block, mean should be 100
# # noise : compute standard deviation of errts
# ktrs=$(1dcat out.keep_trs_rall.txt | tr ' ' ',')
# 3dTstat -mean -prefix rm.signal.all "all_runs.${subj}+orig[${ktrs}]"
# 3dTstat -stdev -prefix rm.noise.all "errts.${subj}_REML+orig[${ktrs}]"
# 3dcalc -a rm.signal.all+orig \
# -b rm.noise.all+orig \
# -expr 'a/b' -prefix TSNR.${subj}
# # --------------------------------------------------
# # compute TSNR stats for dset labels: brain
# compute_ROI_stats.tcsh \
# -out_dir tsnr_stats_regress \
# -stats_file tsnr_stats_regress/stats_auto_brain.txt \
# -dset_ROI mask_epi_anat.${subj}+orig \
# -dset_data TSNR.${subj}+orig \
# -rset_label brain \
# -rval_list 1
# # ---------------------------------------------------
# # compute and store GCOR (global correlation average)
# # (sum of squares of global mean of unit errts)
# 3dTnorm -norm2 -prefix rm.errts.unit errts.${subj}_REML+orig
# 3dmaskave -quiet -mask mask_epi_anat.${subj}+orig \
# rm.errts.unit+orig > mean.errts.unit.1D
# 3dTstat -sos -prefix - mean.errts.unit.1D\' > out.gcor.1D
# echo "-- GCOR = $(cat out.gcor.1D)"
# # ---------------------------------------------------
# # compute correlation volume
# # (per voxel: correlation with masked brain average)
# 3dmaskave -quiet -mask mask_epi_anat.${subj}+orig \
# errts.${subj}_REML+orig > mean.errts.1D
# 3dTcorr1D -prefix corr_brain errts.${subj}_REML+orig mean.errts.1D
# # create ideal files for fixed response stim types
# 1dcat X.nocensor.xmat.1D'[8]' > ideal_ON_H1_R1.1D
# 1dcat X.nocensor.xmat.1D'[9]' > ideal_ON_H2_R1.1D
# 1dcat X.nocensor.xmat.1D'[10]' > ideal_ON_Q1_R2.1D
# 1dcat X.nocensor.xmat.1D'[11]' > ideal_ON_Q2_R2.1D
# 1dcat X.nocensor.xmat.1D'[12]' > ideal_ON_Q3_R2.1D
# 1dcat X.nocensor.xmat.1D'[13]' > ideal_ON_Q4_R2.1D
# # --------------------------------------------------
# # extract non-baseline regressors from the X-matrix,
# # then compute their sum
# 1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
# 3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
# # ============================ blur estimation =============================
# # compute blur estimates
# touch blur_est.${subj}.1D # start with empty file
# # create directory for ACF curve files
# mkdir -p files_ACF
# # -- estimate blur for each run in epits --
# touch blur.epits.1D
# # restrict to uncensored TRs, per run
# for run in 01 02; do
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# if [[ -z "$trs" ]]; then continue; fi
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.epits.r${run}.1D \
# "all_runs.${subj}+orig[${trs}]" >> blur.epits.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\')
# echo "average epits FWHM blurs: $blurs"
# echo "$blurs # epits FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\')
# echo "average epits ACF blurs: $blurs"
# echo "$blurs # epits ACF blur estimates" >> blur_est.${subj}.1D
# # -- estimate blur for each run in errts --
# touch blur.errts.1D
# for run in 01 02; do
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# if [[ -z "$trs" ]]; then continue; fi
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.errts.r${run}.1D \
# "errts.${subj}+orig[${trs}]" >> blur.errts.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\')
# echo "average errts FWHM blurs: $blurs"
# echo "$blurs # errts FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\')
# echo "average errts ACF blurs: $blurs"
# echo "$blurs # errts ACF blur estimates" >> blur_est.${subj}.1D
# # -- estimate blur for each run in err_reml --
# touch blur.err_reml.1D
# for run in 01 02; do
# trs_count=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run | wc -w)
# if [[ $trs_count -eq 0 ]]; then continue; fi
# trs=$(1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run $run)
# 3dFWHMx -detrend -mask mask_epi_anat.${subj}+orig \
# -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r${run}.1D \
# "errts.${subj}_REML+orig[${trs}]" >> blur.err_reml.1D
# done
# # compute average FWHM blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\')
# echo "average err_reml FWHM blurs: $blurs"
# echo "$blurs # err_reml FWHM blur estimates" >> blur_est.${subj}.1D
# # compute average ACF blur (from every other row) and append
# blurs=$(3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\')
# echo "average err_reml ACF blurs: $blurs"
# echo "$blurs # err_reml ACF blur estimates" >> blur_est.${subj}.1D
# # add 3dClustSim results as attributes to any stats dset
# mkdir -p files_ClustSim
# # Get the last ACF line and the 3 lines before it
# acf_comment_line=$(grep -n "err_reml ACF blur estimates" blur_est.${subj}.1D | tail -1 | cut -d: -f1)
# if [[ -n "$acf_comment_line" ]]; then
# # Get the 4 lines ending with that comment (the comment line and 3 lines before it)
# acf_data=$(sed -n "$((acf_comment_line-3)),$acf_comment_line p" blur_est.${subj}.1D)
# echo "ACF data block:"
# echo "$acf_data"
# # Extract just the numbers from those 4 lines, take the first 3
# params=($(echo "$acf_data" | grep -oE '[0-9]+\.[0-9]+' | head -3))
# echo "Extracted params: '${params[0]}' '${params[1]}' '${params[2]}'"
# # Verify we have 3 parameters and first is between 0 and 1
# if [[ ${#params[@]} -eq 3 ]] && (( $(echo "${params[0]} > 0 && ${params[0]} < 1" | bc -l) )); then
# echo "ACF parameters are valid: a=${params[0]}, b=${params[1]}, c=${params[2]}"
# 3dClustSim -both -mask mask_epi_anat.${subj}+orig -acf ${params[0]} ${params[1]} ${params[2]} \
# -cmd 3dClustSim.ACF.cmd -prefix files_ClustSim/ClustSim.ACF
# else
# echo "ERROR: Invalid ACF parameters"
# exit 1
# fi
# else
# echo "ERROR: Could not find err_reml ACF blur estimates line"
# exit 1
# fi
# # Fix the 3drefit command
# if [[ -f 3dClustSim.ACF.cmd ]]; then
# echo "Running 3drefit command:"
# cat 3dClustSim.ACF.cmd
# # Execute the command from the file, adding the dataset names
# cmd_line=$(cat 3dClustSim.ACF.cmd)
# $cmd_line stats.${subj}+orig stats.${subj}_REML+orig
# else
# echo "3dClustSim.ACF.cmd not found, skipping 3drefit step"
# fi
# # ========================= auto block: QC_review ==========================
# # generate quality control review scripts and HTML report
# # generate a review script for the unprocessed EPI data
# # (all echoes of all runs)
# gen_epi_review.py -script @epi_review.${subj} \
# -dsets pb00.${subj}.r*.e*.tcat+orig.HEAD
# # -------------------------------------------------
# # generate scripts to review single subject results
# # (try with defaults, but do not allow bad exit status)
# # write AP uvars into a simple txt file
# cat << EOF > out.ap_uvars.txt
# mot_limit : 0.5
# out_limit : 0.05
# copy_anat : MPRAGE+orig.HEAD
# combine_method : m_tedana
# mask_dset : mask_epi_anat.${subj}+orig.HEAD
# ss_review_dset : out.ss_review.${subj}.txt
# echo_times : 12.3 26.0 40.0
# max_4095_warn_dset : out.4095_warn.txt
# reg_echo : 2
# vlines_tcat_dir : vlines.pb00.tcat
# EOF
# # and convert the txt format to JSON
# cat out.ap_uvars.txt | afni_python_wrapper.py -eval "data_file_to_json()" \
# > out.ap_uvars.json
# # initialize gen_ss_review_scripts.py with out.ap_uvars.json
# gen_ss_review_scripts.py -exit0 \
# -init_uvars_json out.ap_uvars.json \
# -write_uvars_json out.ss_review_uvars.json
# # ========================== auto block: finalize ==========================
# # remove temporary files
# rm -f rm.*
# # --------------------------------------------------
# # if the basic subject review script is here, run it
# # (want this to be the last text output)
# if [[ -e @ss_review_basic ]]; then
# ./@ss_review_basic 2>&1 | tee out.ss_review.${subj}.txt
# # generate html ss review pages
# # (akin to static images from running @ss_review_driver)
# apqc_make_tcsh.py -review_style pythonic -subj_dir . \
# -uvar_json out.ss_review_uvars.json
# apqc_make_html.py -qc_dir QC_${subj}
# echo -e "\nconsider running: \n"
# echo " afni_open -b ${subj}_oldPI_native.results/QC_${subj}/index.html"
# echo ""
# fi
# } 2>&1 | tee "/home/shared/aclab-fmri/Studies/33_MOTIP2018/MRI_data/PREPROCESSED/results/output.proc.${subj}_oldPI_native"
# # return to parent directory (just in case...)
# cd ${preproc_results}
# echo "execution finished: `date`"
# fi
# done
Version:
Precompiled binary linux_openmp_64: Mar 27 2025 (Version AFNI_25.0.11 ‘Severus Alexander’)
Environment (Docker, Singularity / Apptainer, custom installation):
AFNI installed as per standard instructions on Rocky Linux 9.
Relevant log outputs (up to 20 lines):
Due to the character limit I was not able to post the full output, but it contained no concerning errors.
Screenshots / relevant information:
Due to limits, I can’t post the correct-looking dataset, but it just looks… normal.
Below is the “same” stats bucket dataset after my attempt at a “final” pipeline. As you can see, something is very wrong. Again, this is the exact same type of artifact that I saw the very first time I ran this pipeline (start to finish, the proper way, without redoing any part of it).