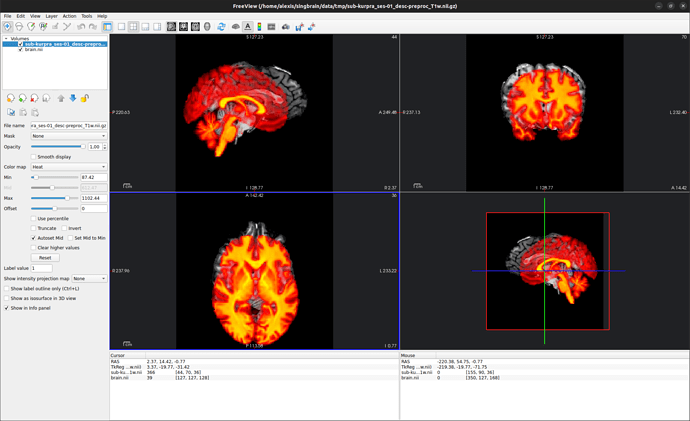
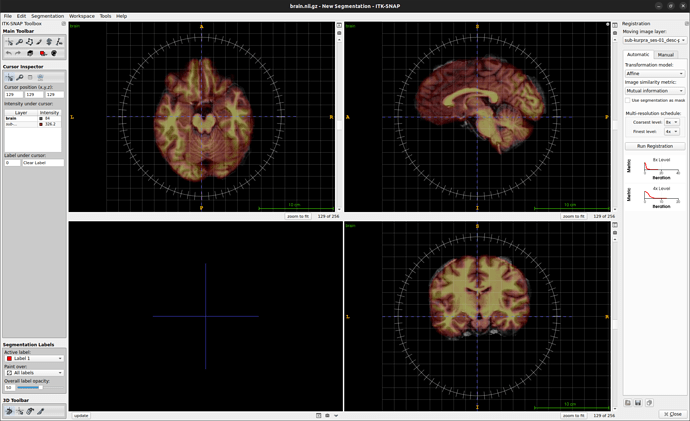
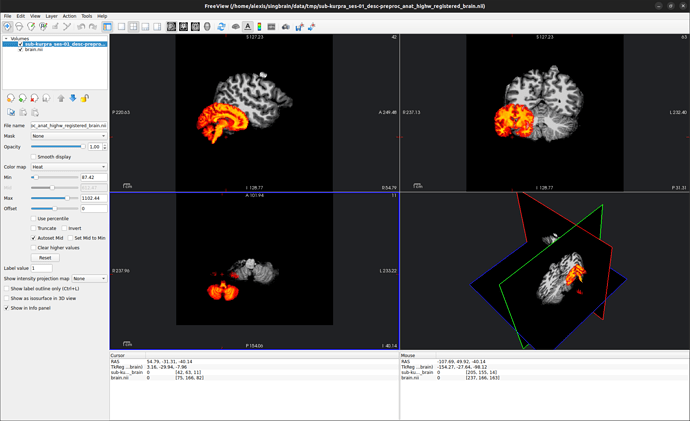
I am trying to register an T1 anatomy IMG2 acquired at the beginning of an fMRI session with a high-resolution T1 anatomy IMG1 that I used with freesurfer’s reconall to extract surfaces.
I went through the following steps:
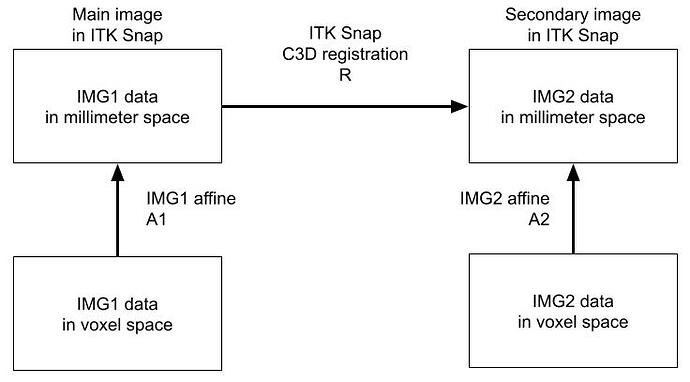
- registered IMG2 on IMG1 in itk-snap (see image B)
- exported computed registration in itk format
- transformed itk registration using
c3d_affine_tool(see command C below), which yielded the following file:
〉 cat registration.mat
0.965521 0.0573185 0.0413778 29.7438
0.0347123 -0.0362805 -0.867828 176.751
-0.0535802 0.973744 -0.00314146 23.563
0 0 0 1
Finally, I applied the computed registration using the following script in Python:
import nibabel as nib
import numpy as np
anat_high_img = nib.load("img1.nii.gz")
anat_sess_img = nib.load("img2.nii.gz")
registration = np.loadtxt("registration.mat", delimiter=" ")
full_registration = anat_high_img.affine @ registration
registered_anat_sess_img = nib.nifti1.Nifti1Image(
anat_sess_img.get_fdata(),
full_registration
)
registered_anat_sess_img.to_filename("img2_registered.nii")
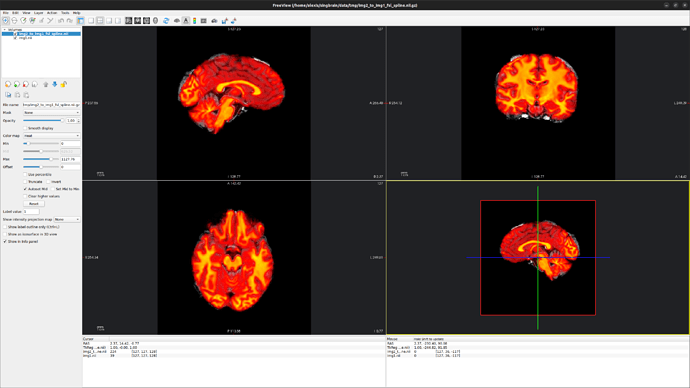
but the image saved is not coregistered to the initial T1 image IMG1 ![]() (see picture D below).
(see picture D below).
I am surprised about this because I used this procedure previously and it had given me co-registered images. I tried a lot of combinations of the registration matrix and the affine functions (using that of anat_sess_img as well, in formulas like full_registration = anat_high_img.affine @ registration @ anat_sess_img.affine), but nothing yielded co-registered images.
Am I missing something here?
What is ITK-snap computing exactly?
Thanks a lot in advance for your help! ![]()
A. IMG1 (grayscale colormap) and IMG2 (heat colormap) in freeview
B. Registring IMG1 on IMG2 in itk-snap
C. c3d_affine_tool command
c3d_affine_tool -ref img1.nii.gz -src img2.nii.gz -itk registration.txt -ras2fsl -o registration.mat
D. Co-registered anatomies are mis-aligned