Hello everyone,
I am a graduate student who is relatively new to neuroscience and am using fmriPrep and AFNI for my processing and analyses. I ran my data through fmriPrep with no errors, and I got all the way to first levels before I noticed that a participant’s brain was much larger than the underlay I was using (MNI 152). I noticed that all my participants were like this, and so all of my results appear as if they are outside of MNI space! Anyone mind helping me figure out the best approach to this? In case it helps, I pasted the 3dInfo output for the fmriPrep-processed bold below:
Dataset File: sub-001_task-framing_run-1_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
Identifier Code: NII_3oWF8omGHyiSncZWxeEJvg Creation Date: Wed Mar 4 15:41:52 2020
Template Space: TLRC
Dataset Type: Echo Planar (-epan)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 379,979,600 (380 million) bytes
Geometry String: “MATRIX(-3,0,0,96,0,-3,0,132,0,0,3,-78):65,77,65”
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -96.000 [R] -to- 96.000 [L] -step- 3.000 mm [ 65 voxels]
A-to-P extent: -96.000 [A] -to- 132.000 [P] -step- 3.000 mm [ 77 voxels]
I-to-S extent: -78.000 [I] -to- 114.000 [S] -step- 3.000 mm [ 65 voxels]
Number of time steps = 292 Time step = 2.00000s Origin = 0.00000s
I appreciate anyone helping out a neuro newbie!
-D
Does this effect appear in fMRIPrep’s reports?
Thanks for the response! Just to make sure I’m understanding you right, are you referring to the HTML files? If so, everything looks fine; nothing looks like it is overly big, and there aren’t any errors…
Do you mean the T1 structure image looks bigger, or the fMRI image looks bigger? If it is the latter, I suspect it is because there is (possibly) CSF above the cortical surface. It usually appears quite bright.
The functional image is the main issue. I use the MNI template as an underlay and whether using first-level or second-level results as an overlay. For my data, I would expect that a good amount of results are cortical; however, when I threshold my results, everything that survives is outside the brain, in coordinates that make little sense. I attached 2 pictures: 1. My data overlaying the template (obviously with little to no thresholding to help illustrate my confusion) and 2. the template overlaying my data (to illustrate that my conundrum doesn’t appear to be due to space around my subjects’ brains, or something similar). Hopefully this helps!

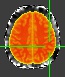
Wow. honestly I have never seen such situation before. The only think I can think of to check is if somehow the T1 images’s resolution is messed up. But that is unlikely. Perhaps some developers of fMRIprep can help?
Well I’m glad to know that I’m not the only one confused! Also, this data was preprocessed a while ago, so I ran a few participants through the pipeline again. I had very slightly different results, but categorically, nothing changed…Since you brought it up, any suggestions on making sure my data is okay? MRIQC and just looking at stuff suggests things are fine…

