Summary of what happened:
Dear experts,
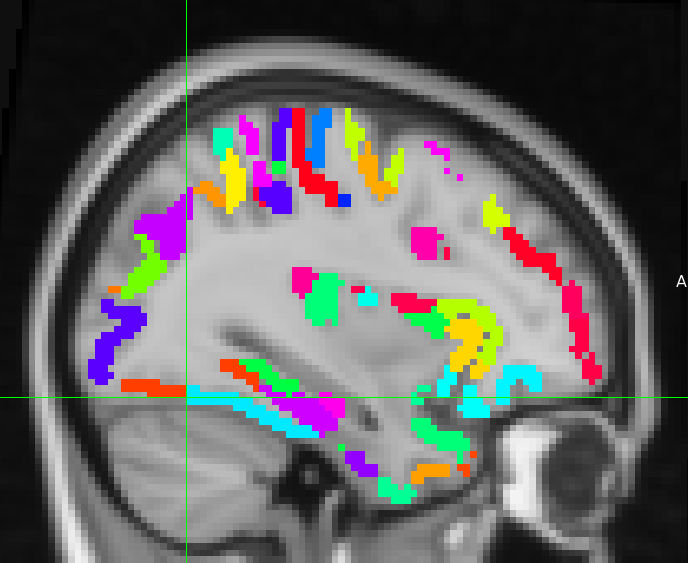
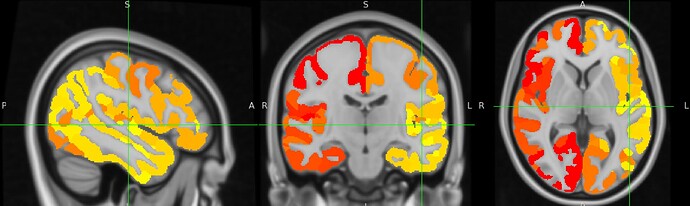
I’m trying to use XCP-D (0.11.1) to denoise my functional images that have been preprocessed using fmrirprep and subsequently extract timeseries using one of the XCP atlases. However, the alignment of the atlases in the xcp_outputfolder/atlases (e.g. atlas-Glasser) - which I assume are being used to extract the timeseries with - is quite poor. I’m confident that this is not an issue with fmriprep because my functional image and the MNI152NLin6Asym_res-2 brain from templateflow overlap perfectly. In fact, the atlases itself do not overlap well with the MNI152NLin6Asym_res-2 template. I see this for both the Glasser and Gordon atlas (see screenshot - I’m only allowed to upload one by the system). What could be the cause of this?
Is it also possible to perform the denoising in T1w space? I tried using a bids filter to select the T1w space images in the func folder of fmriprep but that threw an error.
thanks, Chris
Command used (and if a helper script was used, a link to the helper script or the command generated):
apptainer run \
$XCPD_IMAGE \
${fmriprepdir} ${outputdir} participant \
--input-type fmriprep \
--dummy-scans 3 \
--nuisance-regressors 36P \
--despike \
--file-format nifti \
--participant-label ${subj} \
--work-dir ${workdir} \
--mode linc \
--atlases Glasser
Version:
0.11.1
Environment (Docker, Singularity / Apptainer, custom installation):
apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE
Screenshots / relevant information:
fmriprep output tree
sub-1050
├── anat
│ ├── sub-1050_desc-aparcaseg_dseg.nii.gz
│ ├── sub-1050_desc-aseg_dseg.nii.gz
│ ├── sub-1050_desc-brain_mask.json
│ ├── sub-1050_desc-brain_mask.nii.gz
│ ├── sub-1050_desc-preproc_T1w.json
│ ├── sub-1050_desc-preproc_T1w.nii.gz
│ ├── sub-1050_dseg.nii.gz
│ ├── sub-1050_from-fsnative_to-T1w_mode-image_xfm.txt
│ ├── sub-1050_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5
│ ├── sub-1050_from-MNI152NLin6Asym_to-T1w_mode-image_xfm.h5
│ ├── sub-1050_from-T1w_to-fsnative_mode-image_xfm.txt
│ ├── sub-1050_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5
│ ├── sub-1050_from-T1w_to-MNI152NLin6Asym_mode-image_xfm.h5
│ ├── sub-1050_hemi-L_inflated.surf.gii
│ ├── sub-1050_hemi-L_midthickness.surf.gii
│ ├── sub-1050_hemi-L_pial.surf.gii
│ ├── sub-1050_hemi-L_smoothwm.surf.gii
│ ├── sub-1050_hemi-R_inflated.surf.gii
│ ├── sub-1050_hemi-R_midthickness.surf.gii
│ ├── sub-1050_hemi-R_pial.surf.gii
│ ├── sub-1050_hemi-R_smoothwm.surf.gii
│ ├── sub-1050_label-CSF_probseg.nii.gz
│ ├── sub-1050_label-GM_probseg.nii.gz
│ ├── sub-1050_label-WM_probseg.nii.gz
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_desc-brain_mask.json
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_desc-brain_mask.nii.gz
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_desc-preproc_T1w.json
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_desc-preproc_T1w.nii.gz
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_label-CSF_probseg.nii.gz
│ ├── sub-1050_space-MNI152NLin6Asym_res-2_label-GM_probseg.nii.gz
│ └── sub-1050_space-MNI152NLin6Asym_res-2_label-WM_probseg.nii.gz
├── func
│ ├── aroma_motion_regressors.txt
│ ├── sub-1050_task-rest_AROMAnoiseICs.csv
│ ├── sub-1050_task-rest_desc-brain_mask.json
│ ├── sub-1050_task-rest_desc-brain_mask.nii.gz
│ ├── sub-1050_task-rest_desc-confounds_timeseries.json
│ ├── sub-1050_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-1050_task-rest_desc-MELODIC_mixing.tsv
│ ├── sub-1050_task-rest_desc-preproc_bold.json
│ ├── sub-1050_task-rest_from-scanner_to-T1w_mode-image_xfm.txt
│ ├── sub-1050_task-rest_from-T1w_to-scanner_mode-image_xfm.txt
│ ├── sub-1050_task-rest_space-fsaverage5_hemi-L_bold.func.gii
│ ├── sub-1050_task-rest_space-fsaverage5_hemi-L_bold.json
│ ├── sub-1050_task-rest_space-fsaverage5_hemi-R_bold.func.gii
│ ├── sub-1050_task-rest_space-fsaverage5_hemi-R_bold.json
│ ├── sub-1050_task-rest_space-fsnative_hemi-L_bold.func.gii
│ ├── sub-1050_task-rest_space-fsnative_hemi-L_bold.json
│ ├── sub-1050_task-rest_space-fsnative_hemi-R_bold.func.gii
│ ├── sub-1050_task-rest_space-fsnative_hemi-R_bold.json
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_desc-smoothAROMAnonaggr_bold.json
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_desc-smoothAROMAnonaggr_bold.nii.gz
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_boldref.nii.gz
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-aparcaseg_dseg.nii.gz
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-aseg_dseg.nii.gz
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-brain_mask.json
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-brain_mask.nii.gz
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-preproc_bold.json
│ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-preproc_bold.nii.gz
│ ├── sub-1050_task-rest_space-T1w_boldref.nii.gz
│ ├── sub-1050_task-rest_space-T1w_desc-aparcaseg_dseg.nii.gz
│ ├── sub-1050_task-rest_space-T1w_desc-aseg_dseg.nii.gz
│ ├── sub-1050_task-rest_space-T1w_desc-brain_mask.json
│ ├── sub-1050_task-rest_space-T1w_desc-brain_mask.nii.gz
│ ├── sub-1050_task-rest_space-T1w_desc-preproc_bold.json
│ └── sub-1050_task-rest_space-T1w_desc-preproc_bold.nii.gz
xcp-output tree
atlases
│ ├── atlas-4S1056Parcels
│ │ ├── atlas-4S1056Parcels_dseg.tsv
│ │ ├── atlas-4S1056Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S1056Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S156Parcels
│ │ ├── atlas-4S156Parcels_dseg.tsv
│ │ ├── atlas-4S156Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S156Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S256Parcels
│ │ ├── atlas-4S256Parcels_dseg.tsv
│ │ ├── atlas-4S256Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S256Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S356Parcels
│ │ ├── atlas-4S356Parcels_dseg.tsv
│ │ ├── atlas-4S356Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S356Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S456Parcels
│ │ ├── atlas-4S456Parcels_dseg.tsv
│ │ ├── atlas-4S456Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S456Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S556Parcels
│ │ ├── atlas-4S556Parcels_dseg.tsv
│ │ ├── atlas-4S556Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S556Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S656Parcels
│ │ ├── atlas-4S656Parcels_dseg.tsv
│ │ ├── atlas-4S656Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S656Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S756Parcels
│ │ ├── atlas-4S756Parcels_dseg.tsv
│ │ ├── atlas-4S756Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S756Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S856Parcels
│ │ ├── atlas-4S856Parcels_dseg.tsv
│ │ ├── atlas-4S856Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S856Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-4S956Parcels
│ │ ├── atlas-4S956Parcels_dseg.tsv
│ │ ├── atlas-4S956Parcels_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-4S956Parcels_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-Glasser
│ │ ├── atlas-Glasser_dseg.tsv
│ │ ├── atlas-Glasser_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-Glasser_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-Gordon
│ │ ├── atlas-Gordon_dseg.tsv
│ │ ├── atlas-Gordon_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-Gordon_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-HCP
│ │ ├── atlas-HCP_dseg.tsv
│ │ ├── atlas-HCP_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-HCP_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ ├── atlas-Tian
│ │ ├── atlas-Tian_dseg.tsv
│ │ ├── atlas-Tian_space-MNI152NLin6Asym_res-2_dseg.json
│ │ └── atlas-Tian_space-MNI152NLin6Asym_res-2_dseg.nii.gz
│ └── dataset_description.json
├── dataset_description.json
├── desc-linc_qc.json
├── logs
│ ├── CITATION.bib
│ ├── CITATION.html
│ ├── CITATION.md
│ └── CITATION.tex
├── sub-1050
│ ├── anat
│ │ └── sub-1050_space-MNI152NLin6Asym_desc-preproc_T1w.nii.gz
│ ├── func
│ │ ├── sub-1050_task-rest_desc-preproc_design.json
│ │ ├── sub-1050_task-rest_desc-preproc_design.tsv
│ │ ├── sub-1050_task-rest_motion.json
│ │ ├── sub-1050_task-rest_motion.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_desc-linc_qc.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-denoised_bold.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-denoised_bold.nii.gz
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-denoisedSmoothed_bold.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_desc-denoisedSmoothed_bold.nii.gz
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-alff_boldmap.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-alff_boldmap.nii.gz
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-alff_desc-smooth_boldmap.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-alff_desc-smooth_boldmap.nii.gz
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-reho_boldmap.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_res-2_stat-reho_boldmap.nii.gz
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-alff_bold.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-alff_bold.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-coverage_bold.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-coverage_bold.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-mean_timeseries.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-mean_timeseries.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-pearsoncorrelation_relmat.json
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-pearsoncorrelation_relmat.tsv
│ │ ├── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-reho_bold.json
│ │ └── sub-1050_task-rest_space-MNI152NLin6Asym_seg-Glasser_stat-reho_bold.tsv
├── sub-1050.html