Summary of what happened:
Hi! Thank you very much for all the hard work on making processing neuroimaging data better and more reproducible! This is my first Neurostars post so please bear with me if I’m missing any key information. I’m having trouble getting XCP-D to recognize my fMRIPost-AROMA output, where I’m getting the error “Dataset aroma is not a derivatives dataset. Skipping.”. However, the fMRIPost-AROMA output and dataset_description.json looks fine to me.
Best, Anders
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run -t --rm \
-v ${fmriprep_dir}:/fmri_dir \
-v ${xcp_d_dir}/task-rest:/output_dir \
-v ${work_dir}:/work \
-v /data/Bergen2/BIDS/derivatives/fmripost_aroma/task-rest:/fmripost_aroma \
-v /usr/local/freesurfer/7.4.1-1/license.txt:/opt/freesurfer/license.txt \
-v ${TEMPLATEFLOW_HOME}:/templateflow:ro \
-e TEMPLATEFLOW_HOME=/templateflow \
pennlinc/xcp_d:0.11.0 /fmri_dir /output_dir participant \
--mode linc \
--input-type fmriprep \
--file-format auto \
--work-dir /work \
--nuisance_regressors 'aroma' \
--datasets aroma=/fmripost_aroma \
--participant_label ${subject} \
--notrack \
--verbose \
--smoothing 6 \
--motion-filter-type none \
--fd-thresh 0 \
--output-type auto \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
--min-coverage auto \
--fs-license-file /opt/freesurfer/license.txt \
--nprocs ${nprocs} \
--task-id rest \
--dummy-scans 10 \
> ${xcp_d_dir}/task-rest/logs/${subject}_xcp_d_task-rest.log
Version:
pennlinc/xcp_d:0.11.0
nipreps/fmripost-aroma:0.0.12
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Relevant log outputs (up to 20 lines):
250724-10:08:13,2 nipype.utils IMPORTANT:
Collected run data for sub-001_ses-001_task-rest_space-fsLR_den-91k_bold.dtseries.nii:
boldmask: /fmri_dir/sub-001/ses-001/func/sub-001_ses-001_task-rest_space-MNI152NLin6Asym_res-2_desc-brain_mask.nii.gz
boldref: /fmri_dir/sub-001/ses-001/func/sub-001_ses-001_task-rest_space-MNI152NLin6Asym_res-2_boldref.nii.gz
motion_file: /fmri_dir/sub-001/ses-001/func/sub-001_ses-001_task-rest_desc-confounds_timeseries.tsv
motion_json: /fmri_dir/sub-001/ses-001/func/sub-001_ses-001_task-rest_desc-confounds_timeseries.json
nifti_file: /fmri_dir/sub-001/ses-001/func/sub-001_ses-001_task-rest_space-MNI152NLin6Asym_res-2_desc-preproc_bold.nii.gz
Dataset aroma is not a derivatives dataset. Skipping.
Not required: xcpdatlases
Not required: xcpd4s
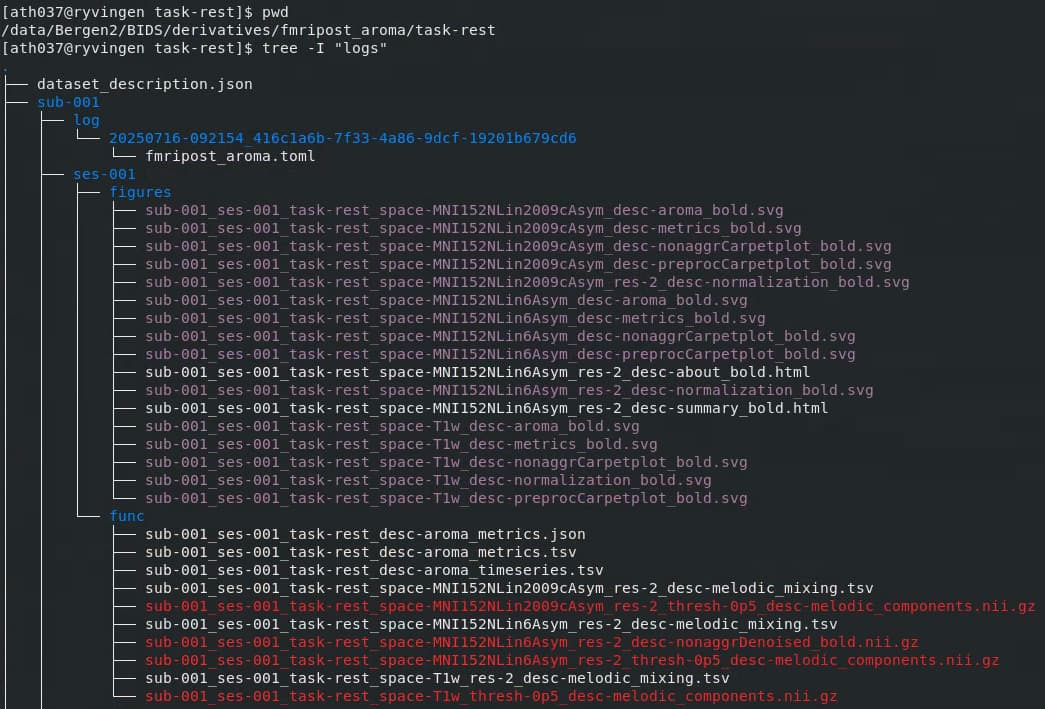
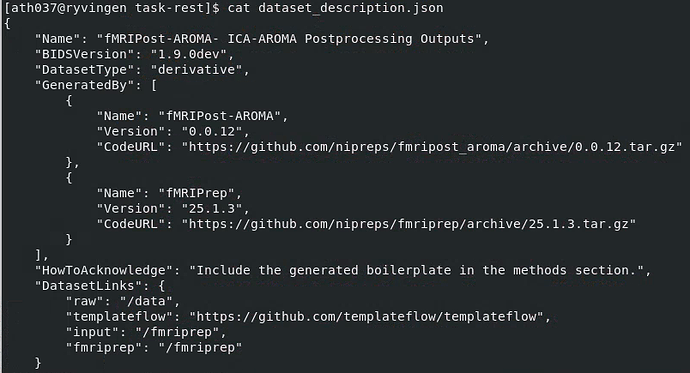
Screenshots / relevant information:
Relevant content of fmripost-aroma output