Hi, I am running Nibabies & XCPD with infant rsfmri data.
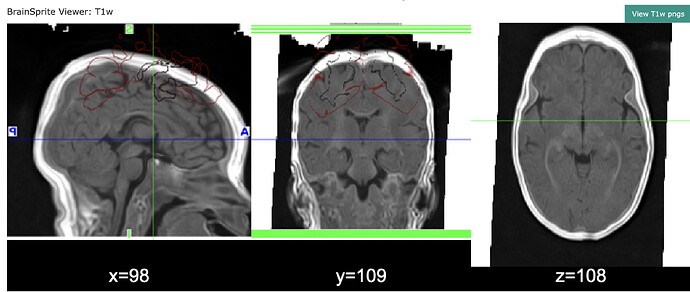
Nibabies outputs look fine, and xcpd looks mostly fine, except for the first Brainsprite images (which I believe are overlaying freesurfer seg onto T1).
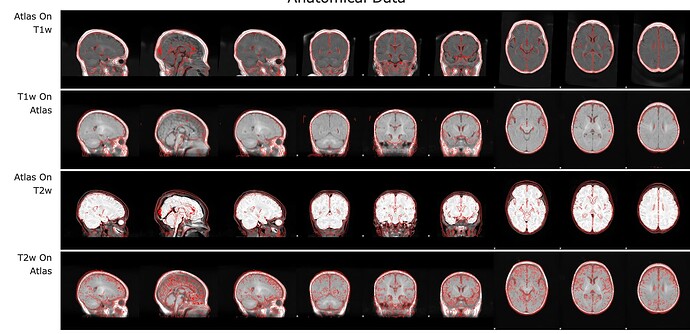
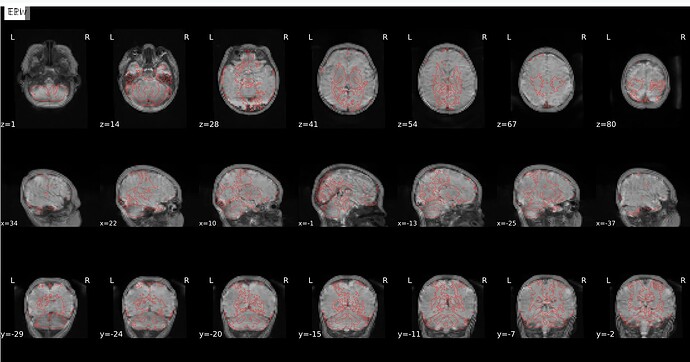
Why is it so off ? and why are other images fine (atlas and anat to func) ?
(I have participant.csv with correct ages setup)
Nibabies Command:
sub-nibabies-25.0.1 -i /MRI_DATA/mritesting/rawdata/ -o /MRI_DATA/mritesting/derivatives/nibabies/v25_0_1 -d /MRI_DATA/mritesting/derivatives/nibabies/work -l /MRI_DATA/mritesting/derivatives/nibabies/v25_0_1/v_25_sub-mc329567_multi.log -a "--participant-label mc329567 --norm-csf --cifti-output 91k"
XCP_D Command:
sub-xcp_d-0.10.0rc3 -i /MRI_DATA/mritesting/derivatives/nibabies/v25_0_1 -o /MRI_DATA/mritesting/derivatives/xcp_d_0_10_0_rc3 -l /MRI_DATA/mritesting/derivatives/xcp_d_0_10_0_rc3/mc329567_xcpd.log -a " --mode hbcd --participant-label mc329567 --min-time 30 --motion-filter-type notch --band-stop-min 15 --band-stop-max 35"