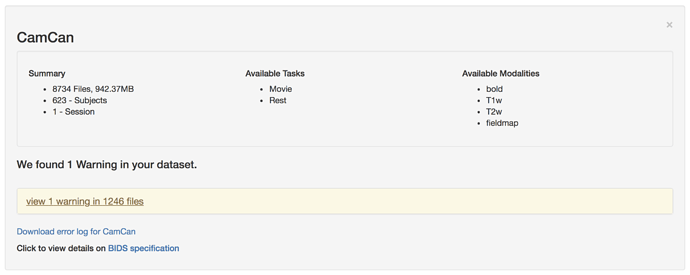
Hi Guys.
Firstly, thanks to everyone who has helped so far.
With regards to the folder structure: we reluctantly separated the modalities because of the enormous quantity of data we are trying to provide to hundreds of researchers. For example, if a researcher requests T1 data, it does not make sense to provide them with all other modalities, which can run into multiple TB for the whole dataset. It is also against our data sharing policy to provide access to data that has not been requested. Do any of you know a way to achieve this without breaking BIDS format?
We plan to remove any reference to BIDS until the issue of how to share separate modalities is resolved, although we will continue to provide data using the same folder structure as before. The benefit is that it is handy to have data gzipped, and the .json file provides another source of information (this has already been useful in identifying the TR issue).
For those who already have the data, here is a brief explanation of the folder structure.
We took the following BIDS folders generated by our aa (automatic analysis) BIDS function:
BIDS/
sub-CC321154/
anat/
dwi/
... and so on
and separated the modalities while maintaining the folder structure.
BIDSsep/
anat/
sub-CC321154/
anat/
dwi/
sub-CC321154/
dwi/
Therefore, recombining the data should have been a simple case of copying those folders back into the original folder structure. If the functions we used to generate the original BIDS folders was working properly, then the result should be BIDS compliant.
If anyone can suggest a better way to do this then would be willing to listen, but remember that we need to grant specific access to each modality, so sharing datasets at the subject level is not an option.
I presume that if BIDS is intended to be a format for sharing data, then this is an important issue to be resolved. It seems like it would be useful to be able to download different modalities / scans from the same subject and combine them at the target location. This would not only allow granular access to large datasets but also aid in sharing data from multi-site studies.
To briefly answer the question of the qform/sform problem: this appears to be a bug in SPM. We are working on a solution with the developers. However, rest assured that as long as the value is >0 then the coordinate system will be numerically correct, but incorrectly labeled as being coregistered to another scan.
Darren