Hi all,
I have a rather specific issue that I’m hoping to get some input on. I got some helpful feedback on Twitter, but Chris (effigies) suggested I post here for a more formal back-and-forth.
I am planning to use fmriprep to analyze a very large existing dataset (Cam-CAN, for those who are familiar). After requesting access, you are granted a server login and can download the data from ~700 participants. Importantly, you get NIFTIs, but not the raw DICOMs.
I had to rework their file directory structure a bit to make it fully BIDS-compatible, but this is what I did (notably, each subject has JSON files in the same directory as the corresponding NIFTI files, but I didn’t see a need to make master JSON files in the parent directory):
CamCan/
dataset_description.json
participants.tsv
each participant folder/
anat/
t1w NIFTIs & JSONs
t2w NIFTIs & JSONs
fmap/
phasediff NIFTIs & JSONs
magnitude 1 NIFTIs (no JSONs)
magnitude 2 NIFTIs (no JSONs)
func/
rest NIFTIs & JSONs
movie NIFTIs & JSONs (BOLD during film viewing)
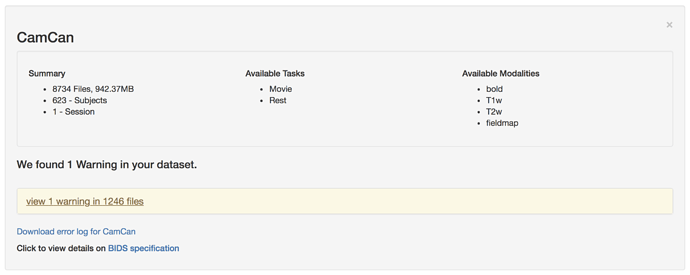
When running the BIDS Validator, I get the following errors:
Error: 3
Repetition time did not match between the scan’s header and the associated JSON metadata file.1246 files
sub-CC110045_task-Movie_bold.nii.gz27256.459 KB | application/x-gzip
Location:
CamCan/sub-CC110045/func/sub-CC110045_task-Movie_bold.nii.gzReason:
Repetition time defined in the JSON (2.47 sec.) did not match the one defined in the NIFTI header (0 sec.)sub-CC110045_task-Rest_bold.nii.gz36890.833 KB | application/x-gzip
Location:
CamCan/sub-CC110045/func/sub-CC110045_task-Rest_bold.nii.gzReason:
Repetition time defined in the JSON (1.97 sec.) did not match the one defined in the NIFTI header (0 sec.)
The TRs being read out from the JSON file (2.47 for movie, 1.97 for rest) are indeed the desired values, but it’s seemingly reading out TR=0 from the NIFTI header. To further complicate matters, this isn’t what the NIFTI header appears to actually contain, if one runs fslhd on a given file, the TR appears to be 1sec (bolded):
fslhd sub-CC723395_task-Movie_bold.nii.gz
filename sub-CC723395_task-Movie_bold.nii.gzsizeof_hdr 348
data_type UINT16
dim0 4
dim1 64
dim2 64
dim3 32
dim4 193
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 512
nbyper 2
bitpix 16
pixdim0 0.000000
pixdim1 3.000000
pixdim2 2.999999
pixdim3 4.440000
pixdim4 1.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.0000
cal_min 0.0000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
time_offset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Aligned Anat
qform_code 2
qto_xyz:1 -2.993574 0.000000 -0.290462 95.590027
qto_xyz:2 0.103233 2.551443 -2.330460 -50.001572
qto_xyz:3 -0.166914 1.578015 3.768048 -120.494217
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Aligned Anat
sform_code 2
sto_xyz:1 -2.993574 0.000000 -0.290466 95.590027
sto_xyz:2 0.103233 2.551443 -2.330460 -50.001572
sto_xyz:3 -0.166917 1.578015 3.768048 -120.494217
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 4D image
aux_file
To confirm this using another tool, running AFNI’s 3dinfo on that same file indicates “Time step = 1.00000s”.
So I have two issues. (1) My NIFTI headers do not contain the proper TR values. Unfortunately, the root of the problem is beyond my control since the data aren’t mine and I’m having to start from these NIFTIs, so I likely need to manually edit the headers. Any input on the optimal way to go about this would be greatly appreciated. (2) The BIDS Validator is nonetheless failing to capture the erroneous TR=1 in the NIFTI header, instead finding TR=0. Thus, I’m concerned that even if I fix the TR values in the NIFTI headers, the BIDS validator (and therefore fmriprep) will probably fail anyway. Should I be concerned with these NIFTIs beyond the TR situation? (If so, that might be a problem of broad interest since a number of publications have been released based on this very dataset…)
Thanks very much in advance.