Summary of what happened:
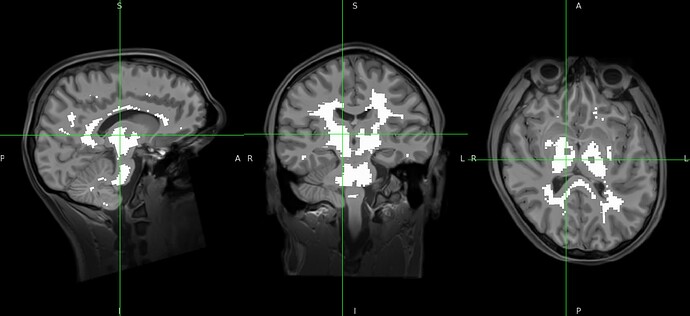
aCompCor mask for white matter includes subcortical structures and may affect subsequent analysis targeting those subcortical structures.
Command used (and if a helper script was used, a link to the helper script or the command generated):
/opt/conda/envs/fmriprep/bin/fmriprep --fs-license-file /work/freesurfer/license.txt /work/PhantomPainBrain /work/PhantomPainBrain/derivatives/fmriprep participant --participant-label AMP08 -w /work/temp_data_PhantomPainBrain --mem-mb 60000 --omp-nthreads 12 --nthreads 16 --longitudinal --ignore slicetiming --fd-spike-threshold 0.5 --dvars-spike-threshold 2.0 --bold2t1w-dof 9 --output-spaces T1w MNI152NLin6Asym:res-2 fsnative fsLR --longitudinal --fs-subjects-dir /work/PhantomPainBrain/derivatives/fmriprep/sourcedata/freesurfer --debug compcor --dummy-scans 0 --derivatives /work/PhantomPainBrain/derivatives/fmriprep --project-goodvoxels --cifti-output 91k --medial-surface-nan
Version:
<23.2.1>
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity/Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS validated dataset
bids-validator@1.14.1
e[33m1: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)e[39m
./README
e[36m Please visit https://neurostars.org/search?q=README_FILE_SMALL for existing conversations about this issue.e[39m
e[34me[4mSummary:e[24me[39m e[34me[4mAvailable Tasks:e[24me[39m e[34me[4mAvailable Modalities:e[39me[24m
4207 Files, 165.03GB VibStim MRI
40 - Subjects rest
4 - Sessions VibStimWrong
Resting
Screenshots / relevant information:
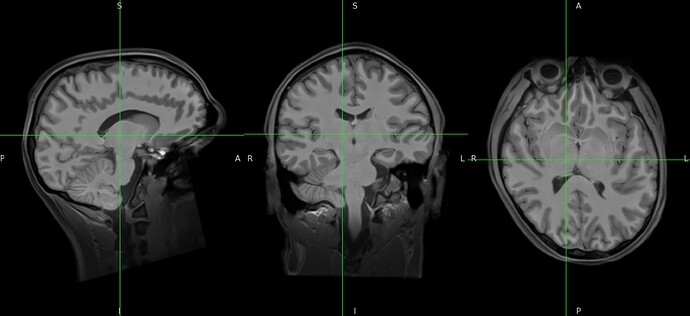
sub-SUB_desc-preproc_T1w.nii.gz
fmriprep_23_2_wf/sub_SUB_wf/bold_ses_BRpost_task_rest_wf/bold_confounds_wf/acc_msk_bin/mapflow/_acc_msk_bin1/acompcor_wm_trans_masked_masked.nii.gz
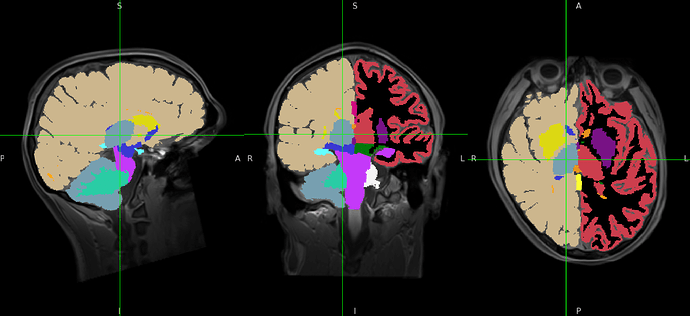
derivatives/fmriprep/sourcedata/freesurfer/sub-SUB/mri/aseg.mgz