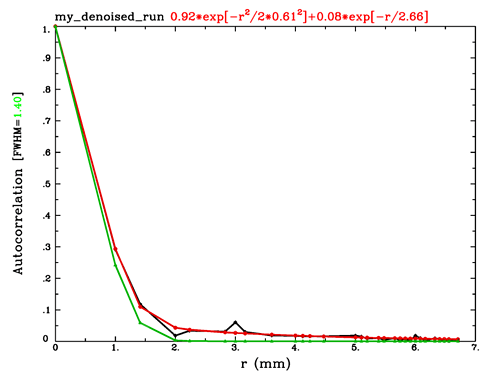
Hi, @meka -
Q1. I would think that it would mostly make sense to calculate the smoothness of residuals, rather than the smoothness of the data itself. When we are talking about noise, that is typically what we are talking about, and that is the smoothness (i.e., spatial autocorrelation function) that is often cared about.
Q3. It is hard to judge the mixed ACF parameters without plotting them. The “show me classic” option
This might be how I would run 3dFWHMx normally—we don’t really care about the Gaussian approx anymore:
3dFWHMx \
-detrend \
-mask mask_epi_anat.FT+tlrc. \
-ACF ACF_FT errts.FT+tlrc. \
> acf_params_FT.txt
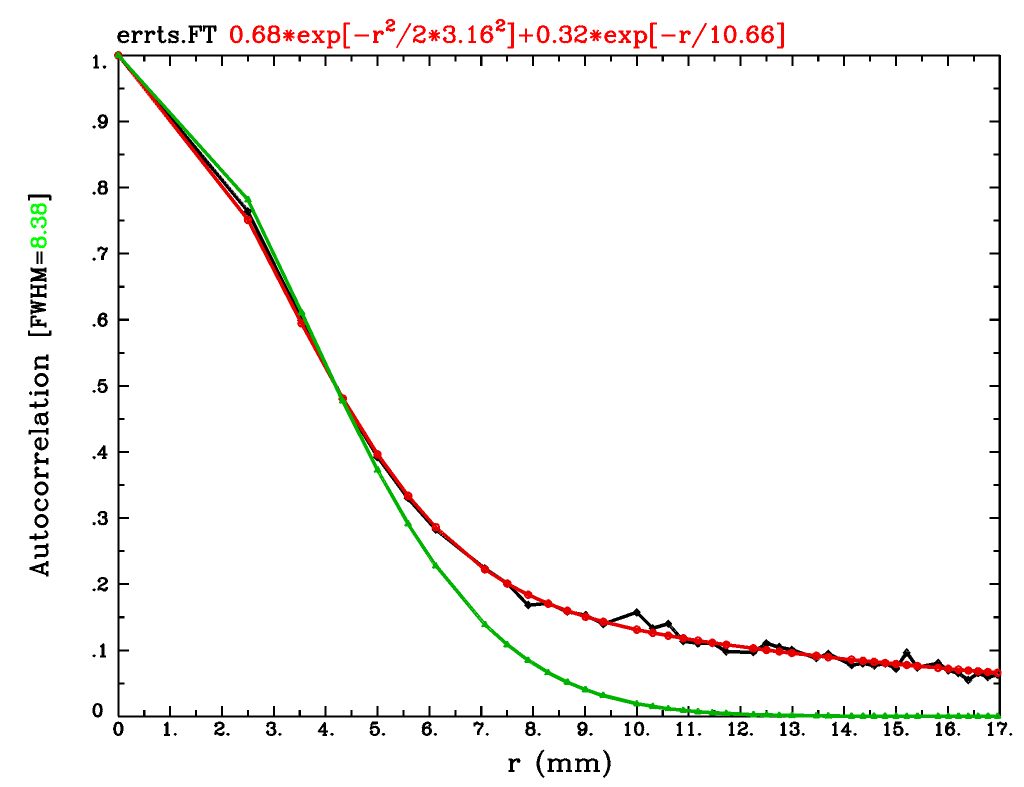
This would generate this text file of params: acf_params_FT.txt
# old-style FWHM parameters
0 0 0 0
# ACF model parameters for a*exp(-r*r/(2*b*b))+(1-a)*exp(-r/c) plus effective FWHM
0.676263 3.1642 10.659 8.38097
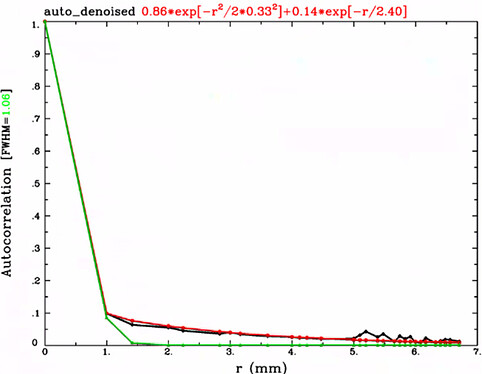
… and this figure of camparing the data points (black) with the new fit (red) and old Gaussian approx (green):
In general, the red mixed-ACF curve will better match the data points, which tend to have a heavier tail than the Gaussian can well approximate (this was a useful observation made in the Eklund et al, 2016 paper about cluster woes—AFNI implemented the mixed ACF model to be a better spatial noise fit, which seems to fit the bill).
Now, if I add the -ShowMeClassicFWHM option in order to also get the Gaussian parameters output
3dFWHMx \
-detrend \
-ShowMeClassicFWHM \
-mask mask_epi_anat.FT+tlrc. \
-ACF ACF_FT_W_GAUSS errts.FT+tlrc. \
> acf_params_FT_w_gauss.txt
… then the exact same figure gets made, and the text file acf_params_FT_w_gauss.txt would look like:
# old-style FWHM parameters
6.13784 5.89771 5.1476 5.71173
# ACF model parameters for a*exp(-r*r/(2*b*b))+(1-a)*exp(-r/c) plus effective FWHM
0.676265 3.16419 10.6591 8.38095
The “old-style” parameter values are not the a,b,c ones of the mixed ACF fit, and generally have very different values (they are basically the FWHM values along each axis, with the last being the “combined” or average).
So, the two sets of numbers you presented both look like “new” mixed ACF parameters, a,b,c. Here is what they look like when I plot them, calling them ‘acf1’ and ‘acf2’, respective to the order you posted them (see below for code I used):
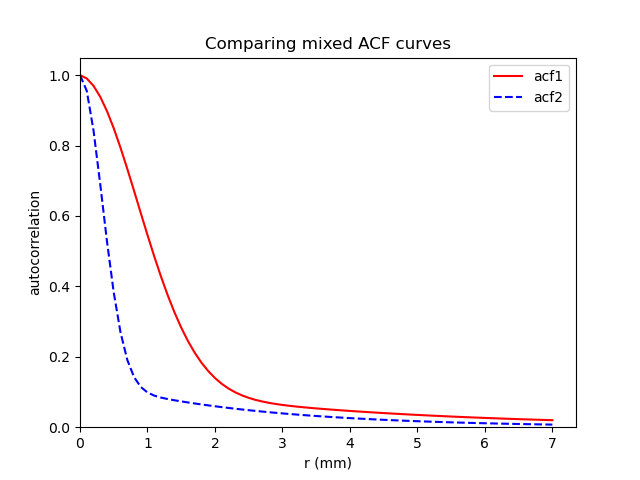
So, I am a bit confused about the two sets of parameters you are setting out—those don’t appear to me to be “classic” (=Gaussian) vs mixed-ACF values—those look much more like two sets of mixed-ACF parameters. I guess it is possible that with veeery anisotropic voxels, your “classic” parameters will be very anisotropic, as well, but I would have thought the smoothness would be larger than each voxel dimension. Your plot looks like what 3dFWHMx would give showing 1 mixed-ACF curve (red) and one Gaussian fit (green), with the data having a fatter tail than the Gaussian can manage and the ACF-fitting it well.
Therefore:
Q4. Could you please verify that the numbers you are providing are really a classic/Gaussian output and mixed-ACF fit?
–pt
Example code for plotting multiple mixed ACF profiles, plot_mixed_acf.py. Edit/add to this acf1 and acf2 variables to plot new/more curves, and execute with python plot_mixed_acf.py from a terminal, or run plot_mixed_acf.py from within IPython or Jupyter-notebook, say.
import numpy as np
import matplotlib.pyplot as plt
def make_default_r_array():
delr = 0.1
return np.arange(0, 7.0+delr, delr)
def mixed_acf(a=None, b=None, c=None, r=None):
"""Calculate an array for displaying the mixed autocorrelation
function (ACF) smoothness estimation parameters from AFNI's 3dFWHMx,
following the empirical relation used/described here:
* Cox RW, Chen G, Glen DR, Reynolds RC, Taylor PA (2017). fMRI
clustering and false-positive rates. Proc Natl Acad Sci
USA. 114(17):E3370-E3371. doi:10.1073/pnas.1614961114
https://pubmed.ncbi.nlm.nih.gov/28420798/
* Cox RW, Chen G, Glen DR, Reynolds RC, Taylor PA (2017). FMRI
Clustering in AFNI: False-Positive Rates Redux. Brain Connect
7(3):152-171. doi: 10.1089/brain.2016.0475.
https://pubmed.ncbi.nlm.nih.gov/28398812/
https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/programs/3dFWHMx_sphx.html#ahelp-3dfwhmx
Parameters
----------
a, b, c : (each a float) parameter values
r : (array of floats) the abscissa values for plotting; default
is [0, 7.0] with step=0.1.
Return
------
y : (array of float) the ordinate values, showing the
mixed ACF curve; length matches that of array r.
"""
BAD_RETURN = np.ndarray([])
# check valid inputs
if a==None or b==None or c==None:
print("** Need to provide values of all 3 input params: a, b, c")
return BAD_RETURN
if hasattr(r, "__len__") :
r = np.array(r)
elif r == None:
r = make_default_r_array()
else:
print("** Need to provide an array of values for r")
return BAD_RETURN
# the actual work
y = a*np.exp(-r*r/(2*b*b))+(1-a)*np.exp(-r/c)
return y
if __name__ == "__main__" :
# example ACF parameter sets, output by 3dFWHMx
acf1 = [0.857931, 0.863189, 3.56048, 1.38152]
acf2 = [0.863087, 0.328694, 2.3981, 1.0597]
# make array of ACF lines
r = make_default_r_array()
y_acf1 = mixed_acf(a=acf1[0], b=acf1[1], c=acf1[2], r=r)
y_acf2 = mixed_acf(a=acf2[0], b=acf2[1], c=acf2[2], r=r)
# make plot
fig1 = plt.figure( "mixed acf plot" )
ax = fig1.add_subplot(111)
ax.plot(r, y_acf1, 'r-', label='acf1')
ax.plot(r, y_acf2, 'b--', label='acf2')
ax.set_xlabel("r (mm)")
ax.set_ylabel("autocorrelation")
ax.set_title("Comparing mixed ACF curves")
ax.set_xlim(left=0)
ax.set_ylim(bottom=0)
ax.legend()
plt.ion()
plt.show()