Summary of what happened:
Our scanner (GE Discovery MR750) was upgraded several months ago, and now instead of a custom multi-echo pulse sequence made by our former MR physicist, we have a GE-made hyperband multi-echo work-in-progress sequence. When I denoise my old data with tedana (0.0.12), it works great. When I denoise my new data from the very same task (same TR, voxel size is smaller due to hyperband), virtually all components are rejected every time, even those that are obviously BOLD. The same is true for a colleague of mine, collecting data from a totally different task - obvious BOLD components are rejected.
The rho/kappa values look almost “reversed”. We initially assumed that the DICOM headers were causing this problem, but after looking them over many times we can’t find the source of the issue (see table below). We are using dcm2niix to convert to NIFTI and catenate the echoes. Looking at the raw echoes, they seem to be as expected (see screenshot below). We have checked over the echo times many times. All the analysis code is the same as before and it works beautifully on data from this same task gathered before the pulse sequence upgrade. Obviously something in the data format is causing this problem but we just can’t think what it might be.
| DV24 (old sequence) | DV29 (ME-WIP, new sequence) |
|---|---|
| 72 x 72 | 128 x 128 |
| 3 mm x 3 mm x 3 mm | 1.7 mm x 1.7 mm x 2 mm |
| No hyperband | Hyperband |
| 13.6 ms / 12.3 ms min echo | 14, 27, 40 ms |
| Pixel Bandwidth: 6944.44 | Pixel Bandwidth: 3906.25 |
| FOV 216 mm | FOV 215.0016 mm |
Command used (and if a helper script was used, a link to the helper script or the command generated):
afni_proc.py \
-subj_id ${sub} \
-script ${preproc_output}/proc.${sub}_${task}_${run} \
-out_dir ${sub}_${task}_${run}.results \
-copy_anat ${data_folder}/HUMAN_EBR-${sub}/${anat_scan_num}/MPRAGE.nii \
-anat_has_skull yes \
-dsets_me_run ${data_folder}/HUMAN_EBR-${sub}/${scan_num}/*e*.nii \
-echo_times 14 27 40 \
-blocks despike tshift align tlrc volreg mask combine scale regress \
-radial_correlate_blocks tcat volreg \
-tcat_remove_first_trs 2 \
-tlrc_base ${data_folder}/MNI_caez_N27+tlrc.BRIK \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-mask_epi_anat yes \
-combine_method m_tedana \
-html_review_style pythonic
tedana -d pb03.$subj.r$run.e*.volreg+tlrc.HEAD
-e $echo_times
–mask mask_epi_anat.$subj+tlrc.HEAD
–out-dir tedana_r$run --convention orig
Version:
0.0.12
Environment (Docker, Singularity, custom installation):
tedana is called from within AFNI by various bash scripts, and is generally working fine for data that is not generated by our new sequence
Relevant log outputs (up to 20 lines):
(The last of many re-attempts at ICA)
INFO ica:tedica:85 ICA with random seed 51 converged in 47 iterations
INFO tedana:tedana_workflow:671 Making second component selection guess from ICA results
INFO collect:generate_metrics:123 Calculating weight maps
INFO collect:generate_metrics:132 Calculating parameter estimate maps for optimally combined data
INFO collect:generate_metrics:145 Calculating z-statistic maps
INFO collect:generate_metrics:155 Calculating F-statistic maps
INFO collect:generate_metrics:165 Thresholding z-statistic maps
INFO collect:generate_metrics:172 Calculating T2* F-statistic maps
INFO collect:generate_metrics:179 Calculating S0 F-statistic maps
INFO collect:generate_metrics:187 Counting significant voxels in T2* F-statistic maps
INFO collect:generate_metrics:193 Counting significant voxels in S0 F-statistic maps
INFO collect:generate_metrics:200 Thresholding optimal combination beta maps to match T2* F-statistic maps
INFO collect:generate_metrics:206 Thresholding optimal combination beta maps to match S0 F-statistic maps
INFO collect:generate_metrics:213 Calculating kappa and rho
INFO collect:generate_metrics:222 Calculating variance explained
INFO collect:generate_metrics:228 Calculating normalized variance explained
INFO collect:generate_metrics:236 Calculating DSI between thresholded T2* F-statistic and optimal combination beta maps
INFO collect:generate_metrics:247 Calculating DSI between thresholded S0 F-statistic and optimal combination beta maps
INFO collect:generate_metrics:257 Calculating signal-noise t-statistics
INFO collect:generate_metrics:295 Counting significant noise voxels from z-statistic maps
INFO collect:generate_metrics:306 Calculating decision table score
INFO tedica:kundu_selection_v2:138 Performing ICA component selection with Kundu decision tree v2.5
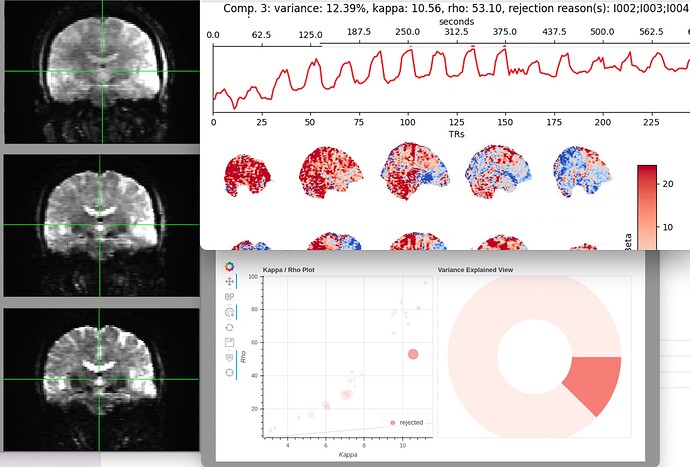
WARNING tedica:kundu_selection_v2:213 No BOLD-like components detected. Ignoring all remaining components.
WARNING tedana:tedana_workflow:701 No BOLD components found, but maximum number of restarts reached.
WARNING tedana:tedana_workflow:773 No BOLD components detected! Please check data and results!
INFO io:denoise_ts:374 Variance explained by decomposition: 59.33%
INFO io:write_split_ts:439 Writing low-Kappa time series: /export/home/shared/aclab-fmri/Studies/33_MOTIP2018/MRI_data/PREPROCESSED/results/512_newPI_2.results/tedana_r01/lowk_ts_OC.nii.gz
INFO io:write_split_ts:446 Writing denoised time series: /export/home/shared/aclab-fmri/Studies/33_MOTIP2018/MRI_data/PREPROCESSED/results/512_newPI_2.results/tedana_r01/dn_ts_OC.nii.gz
INFO io:writeresults:498 Writing full ICA coefficient feature set: /export/home/shared/aclab-fmri/Studies/33_MOTIP2018/MRI_data/PREPROCESSED/results/512_newPI_2.results/tedana_r01/ica_components.nii.gz
INFO tedana:tedana_workflow:889 Making figures folder with static component maps and timecourse plots.
INFO io:denoise_ts:374 Variance explained by decomposition: 59.33%
INFO tedana:tedana_workflow:918 Generating dynamic report
INFO tedana:tedana_workflow:921 Workflow completed
end
Screenshots / relevant information:
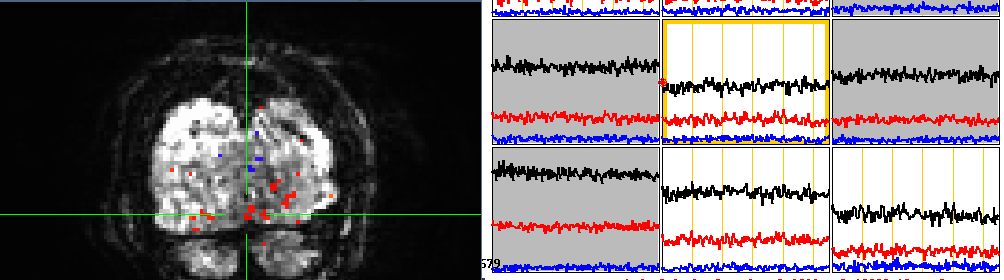
Echoes 1(top), 2(middle), 3(bottom) in raw data converted from DICOM to NIFTI using dcm2niix
Rejected component showing BOLD signal from block design task