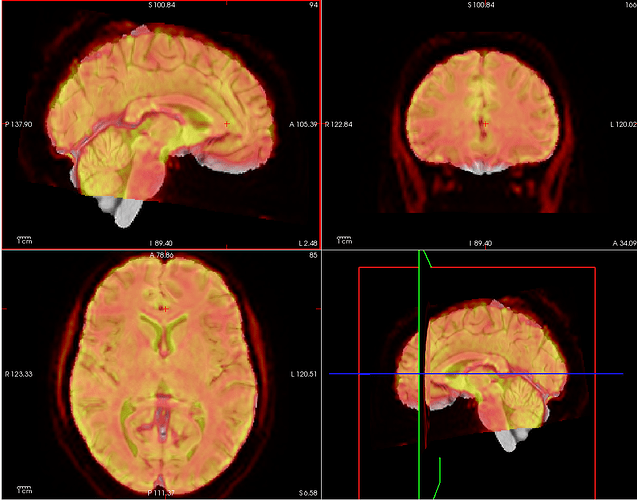
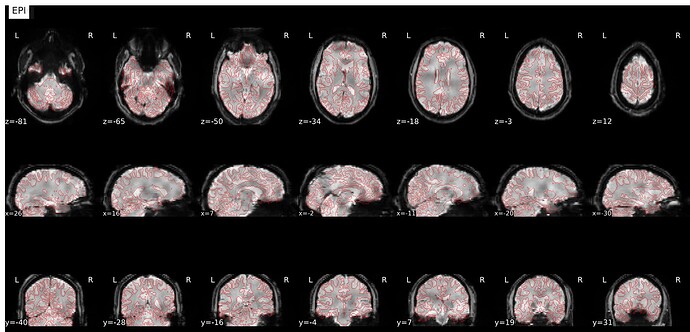
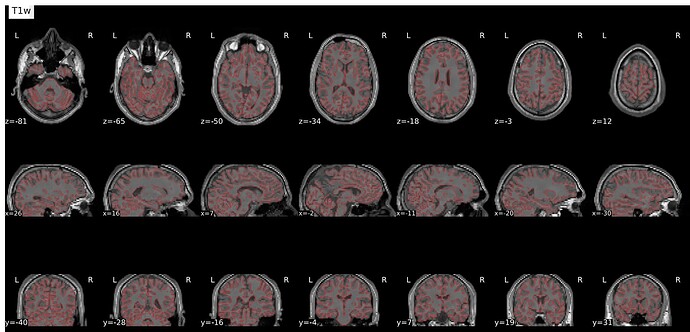
Summary of what happened:
I see a consistent misalignment between the anatomical and functional images generated with fmriprep. (new to fmriprep). I updated the pipeline with some commands I added, but made no difference to the output.
Command used
#!/bin/bash
# User inputs:
bids_root_dir=$HOME/Documents/LUCA/DATA
nthreads=4
mem=25 # in GB
subj=sub-02
# Begin:
echo "Starting the script..."
chmod -R +r /home/maria/Documents/LUCA/DATA/sub-*/func/
# Convert virtual memory from GB to MB
mem=$(echo "${mem//[!0-9]/}") # Remove 'gb' at end
mem_mb=$((mem * 1000 - 5000)) # Reduce some memory for buffer space during pre-processing
echo "Memory allocated: ${mem_mb} MB"
# Set environment variables
export TEMPLATEFLOW_HOME=/home/maria/Documents/LUCA/PREPvalidation/templateflow
export FS_LICENSE=/home/maria/Documents/LUCA/DATA/license.txt
echo "FS License file path: $FS_LICENSE"
# Check if the Docker daemon is running
if ! docker info > /dev/null 2>&1; then
echo "Docker daemon is not running. Please start Docker."
exit 1
else
echo "Docker daemon is running."
fi
echo "Running with Docker..."
docker run --rm -it \
-v /home/maria/Documents/LUCA/DATA:/Luca \
-v /media/maria/Storage-8TB/fmriprep_derivatives:/out \
-v /media/maria/Storage-8TB/BIDSData:/work \
-v /home/maria/Documents/LUCA/DATA/license.txt:/license.txt:ro \
nipreps/fmriprep:24.1.1 \
/Luca /out \
participant \
--bold2t1w-dof 9 \
--bold2t1w-init register \
--participant-label $subj \
--skip-bids-validation \
--fs-license-file /license.txt \
--output-spaces MNI152NLin2009cAsym fsaverage \
--use-aroma true \
--use-syn-sdc \
--force-bbr \
--nthreads $nthreads \
--stop-on-first-crash \
--mem_mb $mem_mb \
-w /work
# Check for errors
if [ $? -ne 0 ]; then
echo "An error occurred while processing participant $subj. Exiting."
exit 1
fi
echo "All participants processed."
Version:
fmriprep 24.1.1
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS validator shows no errors
Relevant log outputs (up to 20 lines):
No errors to report.
Screenshots / relevant information:
Overlay was made using freeview, I changed the functional image to a heat map and made it translucent. I couldn’t spot a problem with the html report (which I can’t post as I am a new user) but there are some overlapping issues I don’t know how to solve. I would greatly appreciate any help!